Plants paper notes
Related: 230529-1413 Plants datasets taxonomy
Key info
PlantCLEF
- PlantCLEF 2021 and 2022 summary papers, no doi :(
- Latest datasets not available, previous ones use eol and therefore are a mix of stuff
- Tasks and datasets differ by year (=can’t reliably do baseline), and main ideas differ too:
- 2021: use Herbaria to supplement lacking real-life photos
- Best methods were the ones that used domain-specific adaptations as opposed to simple CNNs
- 2022: multi-image(/metadata) class. problem with A LOT of classes (80k)
- Classes mean a lot of gimmicks to handle this memory-wise
- 2021: use Herbaria to supplement lacking real-life photos
- Why this doesn’t work for us:
- datasets not available!
- the ones that are are a mix of stuff
- A lot of methods that work there well are specific to the task, as opposed to the general thing
- People can use their own datasets for training
Metrics: MRR (=not comparable to some other literature, even if there were results on the same dataset)
PlantNet300k3 paper
- Dataset is a representative subsample of the big PlantNet dataaset that “covers over 35K species illustrated by nearly 12 million validated images”
- Subset has “306,146 plant images covering 1,081 species.
- Long-tailed distribution of classes:
- 80% of the species account for only 11% of the total number of images”
- Top1 accuracy is OK, but not meaningful
- Macro-average top-1 accuracy differs by A LOT
- The paper does a baselines using a lot of networks
Useful stuff
Citizen science
-
Citizen science (similar to [..] participatory/volunteer monitoring) is scientific research conducted with participation from the general public
most citizen science research publications being in the fields of biology and conservation
-
can mean multiple things, usually using citizens acting volunteers to help monitor/classify/.. stuff (but also citizens initiating stuff; also: educating the public about scientific methods, e.g. schools)
-
allowed users to upload photos of a plant species and its components, enter its characteristics (such as color and size), compare it against a catalog photo and classify it. The classification results are juried by crowdsourced ratings.4
-
Papers
- Paper about using Pl@ntNet5 for CS:
-
“Here we present two Pl@ntNet citizen science initiatives used by conservation practitioners in Europe (France) and Africa (Kenya).”
- paper citing it are interesting: Bonnet: How citizen scientists contribute to monitor… - Google Scholar
- Pl@ntNet can be
- limited for subsets of plants
- limiting plants based on GPS coordinates
- made to train better certain species by manually adding good examples as done in the Lewa Conservatory in Kenya
-
- Assessing accuracy in citizen science-based plant phenology monitoring | SpringerLink <
@fuccilloAssessingAccuracyCitizen2015(2015) z>-
Volunteers demonstrated greatest overall accuracy identifying unfolded leaves, ripe fruits, and open flowers.
- Maybe we’ll want to compare the areas where people are better at than ML in our paper?
-
- Similar to the above, but detecting weeds:Assessing citizen science data quality: an invasive species case study
<
@crallAssessingCitizenScience2011Assessing citizen science data quality (2011) z>- Compare to the paper about detecting weeds with DL: <
@chenPerformanceEvaluationDeep2021(2021) z>
- Compare to the paper about detecting weeds with DL: <
Centralized repositories of stuff
- GBIF (ofc)
- https://bien.nceas.ucsb.edu/bien/ more than 200k observations, and
- This:
- Georeferenced plant observations from herbarium, plot, and trait records;
- Plot inventories and surveys;
- Species geographic distribution maps;
- Plant traits;
- A species-level phylogeny for all plants in the New World;
- Cross-continent, continent, and country-level species lists.
- This:
- No names known to me in their Data contributors
Biodiversity
- Really nice paper: <
@ortizReviewInteractionsBiodiversity2021A review of the interactions between biodiversity, agriculture, climate change, and international trade (2021) z/d> - TL;DR climate change is not the worst wrt biodiversity
Positioning / strategy
Main bits
- Plant classification as a method to monitor biodiversity in the context of citizen science
Why plant classification is hard
- A lot of cleanly labeled herbaria, few labeled pictures (esp. tropical), but trasferring learned stuff from herbarium sheets to field photos is challenging:
-
(e.g. strong colour variation and the transformation of 3D objects after pressing like fruits and flowers) <
@waldchenMachineLearningImage2018(2018) z> - PlantCLEF2021 was entirely dedicated to using herbaria+photos, and there domain adaptations (joint representation space between herb+field) dramatically outperform best classical CNN, esp. the most difficult plants.<
@goeau2021overview(2021) z><@goeauAIbasedIdentificationPlant2021(2021) z>
-
- Connected to the above: lab-based VS field-based investigations
- lab-based has strict protocols for aquisition, people with mobile phones don’t
-
“Lab-based setting is often used by biologist that brings the specimen (e.g. insects or plants) to the lab for inspecting them, to identify them and mostly to archive them. In this setting, the image acquisition can be controlled and standardised. In contrast to field-based investigations, where images of the specimen are taken in-situ without a controllable capturing procedure and system. For fieldbased investigations, typically a mobile device or camera is used for image acquisition and the specimen is alive when taking the picture (Martineau et al., 2017). ”<
@waldchenMachineLearningImage2018(2018) z>
-
- lab-based has strict protocols for aquisition, people with mobile phones don’t
- Phenology (growth stages / seasons -> flowers) make life harder
- Plants sometimes have strong phenology (like bright red flowers) that make it more different and easier to find (esp. here in detecting them in satellite pictures: <
@pearseDeepLearningPhenology2021(2021) z>, but there DL failed less without flowers than non-DL), but sometimes don’t - And ofc. a plant with and without flowers looks like a totally different plant
- Related:
- Plant growth form has been the most helpful species metadata in PlantCLEF2021, but some plants at different stages of growth look like different plant stages.
- Plants sometimes have strong phenology (like bright red flowers) that make it more different and easier to find (esp. here in detecting them in satellite pictures: <
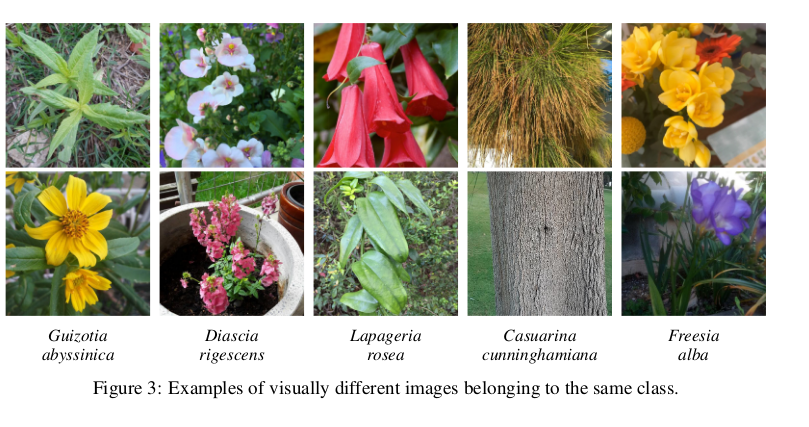
- Intra-species variability
- The Pl@ntNet-300k paper mentions
- epistemic (model) uncertainty (flowers etc.)
- aleatoric (data) uncertainty (small information given to make a decision)
- Plants belonging to the same genus can be visually very similar to each other:
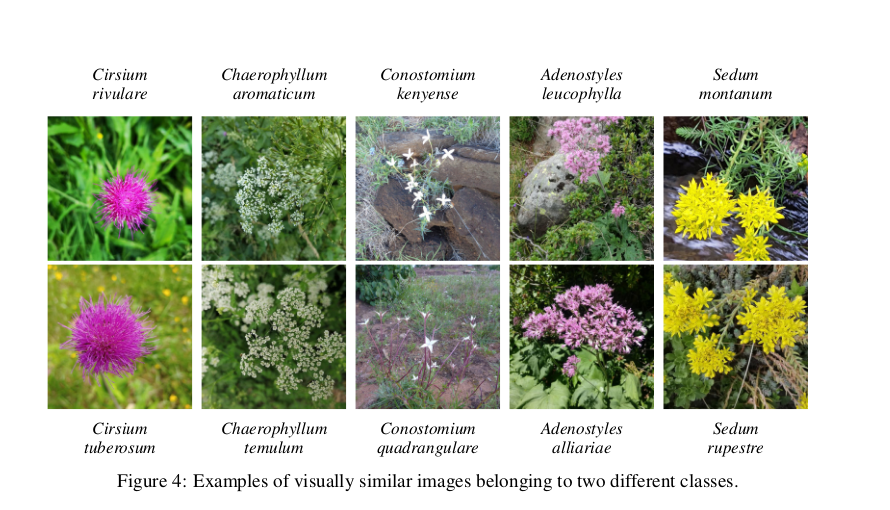 (same paper)
(same paper)
- Plants belonging to the same genus can be visually very similar to each other:
- long-tailed distribution, which: <
@walkerHarnessingLargeScaleHerbarium2022(2022) z/d>- is representative of RL
- is a problem because DL is “data-hungry”
- some say there are a lot of mislabeled specimens <
@goodwinWidespreadMistakenIdentity2015(2015) z/d>
Datasets
EDIT separate post about this: 230529-1413 Plants datasets taxonomy
-
We can classify existing datasets in two types:
- Pl@ntNet / iNaturalist? / …: people with phones
- Clean standardized things like the Plant seedling classification dataset (<
@giselssonPublicImageDatabase2017(2017) z>), common weeds in Denmark dataset <@leminenmadsenOpenPlantPhenotype2020(2020) z/d> etc.- I’d put leaf datasets in this category too
- FloraIncognita is an interesting case:
-
FloraCapture requests contributors to photograph plants from at least five precisely defined perspectives
-
-
There are some special datasets, satellite and whatever, but especially:
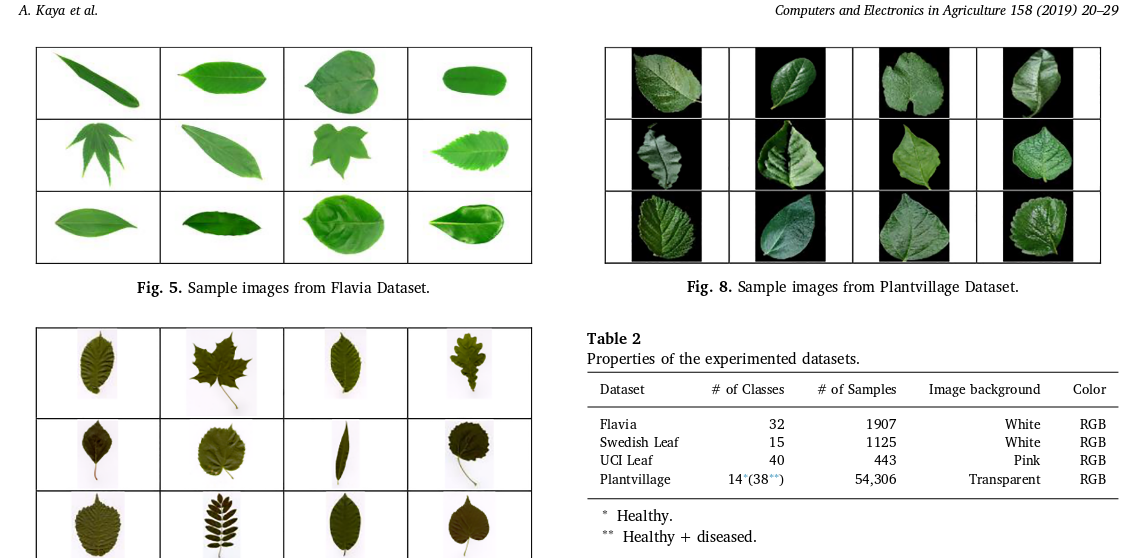
- Leaf datasets exist and are used surprisingly often (if not exclusively) in overviews like the one we want to do:
- Flower datasets / “Natural flower classification”
- Seedlings etc. seem to be useful in industry (and they go hand-in-hand with weed-control)
- Fruit classification/segmentation and other very specific stuff we don’t really care about (<
@mamatAdvancedTechnologyAgriculture2022Advanced Technology in Agriculture Industry by Implementing Image Annotation Technique and Deep Learning Approach (2022) z/d> has an excellent overview of these)
-
Additional info present in datasets or useful:
- PlantCLEF2021 had additional metadata at the species level: growth form, habitat (forest, wetland, ..), and three others
-
- PlantCLEF2022: 3/5 taxonomic levels where used in various ways. Taxonomic loss is a thing (TODO - was this useful?)6
- Pl@ntNet and FloraIncognita apps (can) use GPS coordinates during the decision
- TODO Phenology / phenological stage: is this true to begin with?
Research questions similar to ours
Plant classification (a.k.a. species identification) on pictures
- Things like ecology and habitat loss, citizen science etc.
- Industry:
- Weed detection
Crop identification (sattellites)
Crop stage identification / phenology (sattellites)
Paper outline sketch
Introduction
- Tasks about plants are important
- Ecology: global warming etc., means different distribution of plant species, phenology stages changed, broken balances and stuff and one needs to track it; herbaria and digitization / labeling of herbaria
- Industry: crops stages identification, crops/weeds identification, fruit ripeness identification, etc. long list
- automatic methods have been used, starting from SVM/manual-feature-xxx, later - DL
- DL has been especially nice and improved stuff in all of these different sub-areas, show the examples that compare DL-vs-non-DL in the narrow fields
- The closest relevant thing is PlantCLEF competition that’s really really nice but \textbf{TODO what are we doing that PlantCLEF isn’t?}
- Goal of this paper is:
- Do a short overview of the tasks-connected-to-plants that exist and are usually tackled using AI magic
- Along the way: WHICH AI magic is usually used for the tasks that are formalized as image classification (TODO and object segmentation/detection?)
- Show that while
-
http://ceur-ws.org/Vol-2936/paper-122.pdf / <
@goeau2021overview(2021) z> ↩︎ -
https://hal-lirmm.ccsd.cnrs.fr/lirmm-03793591/file/paper-153.pdf / <
@goeau2022overview(2022) z> ↩︎ -
IBM and SAP open up big data platforms for citizen science | Guardian sustainable business | The Guardian ↩︎
-
Deep Learning with Taxonomic Loss for Plant Identification - PMC ↩︎
 (<
(< (pic from <
(pic from <