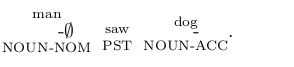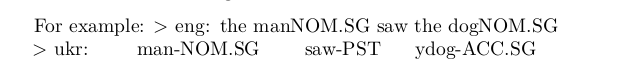serhii.net
In the middle of the desert you can say anything you want
-
Day 1842 (16 Jan 2024)
LaTeX best practices and compression
EDIT: this is becoming a more generic thingy for everything I’d ever need to refer to when writing a paper, TODO clean this at some point.
Resources – DREAM Lab links to https://dream.cs.umass.edu/wp-content/uploads/2020/04/Tips-and-Best-Practices.pdf. Until I set up a system to save PDF info, I’ll paste it as screenshots here:



ChatGPT summarized the relevant pages of the PDF file thus, but didn’t do it well, mostly rewriting myself:
General Formatting
- Smart Quotes: Use `x’ and ``x’’ for single and double quotes respectively.
- Large Numbers: Write large numbers as 54{,}000.
- Word Breaking: Use - to suggest hyphenation points in long words, including ones that already have hyphens inside them. Such words are likely to go over margins etc.:
multi-discipli\-nary - Overflow Lines:
\begin{sloppypar}...for paragraphs where latex goes over the margin. - Float Positioning: Place figures and tables at the top of the page for readability:
\begin{figure}[t] - Center Align Floats: Use
\centeringfor aligning tables and figures.
Specific Elements
- Non-Breaking Space: Use the tilde (~) to keep words together, including always
- before citations:
sth~\cite{whatever} - after numbers in text
- before figure/section references
- before citations:
- Emphasize vs. Bold: Prefer
\emphover bold or\textit.
Equations and Macros
- Macros: Create macros for things that will be changed later, such as DATASET NAMES.
\newcommand{\system}{SQuID\xspace}\xspacehere adds as space unless end of sentence. Package\usepackage{xspace}
Spacing and Sizing
- Paragraph Spacing: Use
\smallskip,\medskip, and\bigskip, instead of\vspace - Line and Text Width: Use fractions of
\linewidthor\textwidth. - Resizing Tables/Figures: Use
\resizeboxwith appropriate dimensions.
Compression hacks
- (see pictures)
- Make spaces smaller around lists:
\begin{itemize} \setlength{\itemsep}{0pt} \setlength{\parskip}{0pt}Paper writing hacks

Even more
Non-breaking spaces
best practices - When should I use non-breaking space? - TeX - LaTeX Stack Exchange lists ALL the places where Knuth wanted people to put nonbreaking spaces, incl:
1)~one 2)~twoDonald~E. Knuth1,~2Chapter~12
Less obvious and not from him:
I~am
Also:
- before all cites, refs
- … and inlined equations
Citations
- ChatGPT says that citations should come before footnotes to prioritize the scholarly source over unimportant info.
- Citations go before the last period of the sentence. EDIT: NOT PARENTHESES, THEY SHOULD BE WITHIN PARENTHESES.4 DAMN
Other bits
- I sometimes write
and around ~50%forgetting that~is a nbsp — hard to catch when reading the text. - As when writing code I like to add some
assert False(or a failing test) so that I know where I stopped the last time,\latexstopcompiling hereis a neat way to make sure I REALLY finis ha certain line I started but not finished.
Latex algorithms bits
Require/ensure
Require/Ensure is basically Input/Output and can be renamed thus1:
\floatname{algorithm}{Procedure} \renewcommand{\algorithmicrequire}{\textbf{Input:}} \renewcommand{\algorithmicensure}{\textbf{Output:}}Sample algpseudocode algorithm float
\usepackage{algorithm} \usepackage{algpseudocode} % ... \begin{algorithm} \caption{Drop Rare Species per Country} \label{alg:drop} \begin{algorithmic} \Require $D_0$: initial set of occurrences \Ensure $D_1$: Set of occurrences after filtering rare species \State $D_1 \gets$ \emptyset \For{each $c$ in Countries} \For{each $s$ in Species} \If {$|O_{c,s} \in D_0| \geq 10$} % if observations of species in country in D_0 have more than 10 entries; || is set cardinality \State{$D_1 \gets D_1 \cup O_{c,s}$} \EndIf \EndFor \EndFor \end{algorithmic} \end{algorithm}
-
Day 1837 (11 Jan 2024)
Yet another jupyter pandas template thing
- Updated
IPython.core.display->IPython.display, removed Black. - See also 250922-1638 jupyter notebook jupyterlab extensions with uv.
- For that setup you’d need
uv add itables
- For that setup you’d need
from pathlib import Path import pandas as pd import seaborn as sns import matplotlib.pyplot as plt INTERACTIVE_TABLES=False # 100% width table from IPython.display import display, HTML display(HTML("<style>.container { width:100% !important; }</style>")) if INTERACTIVE_TABLES: from itables import init_notebook_mode init_notebook_mode(all_interactive=True, connected=True) # column/row limits removal pd.set_option("display.max_columns", None) pd.set_option('display.max_rows', 100) # figsize is figsize plt.rcParams["figure.figsize"] = (6, 8) plt.rcParams["figure.dpi"] = 100 # CHANGEME PATH_STR = "xxxxx/home/sh/hsa/plants/inat500k/gbif.metadata.csv" PATH = Path(PATH_STR) assert PATH.exists()
- Updated
-
Day 1834 (08 Jan 2024)
More adventures plotting geodata
List of all map providers, not all included in geopandas and some paid, nevertheless really neat: https://xyzservices.readthedocs.io/en/stable/gallery.html
-
Day 1830 (04 Jan 2024)
Notes in creating a tale motif ontology for my Masterarbeit
For the 231024-1704 Master thesis task CBT task of my 230928-1745 Masterarbeit draft, I’d like to create an ontology I can use to “seed” LMs to generate ungoogleable stories.
And it’s gonna be fascinating.
I don’t know what’s the difference between knowledge graph, ontology etc. at this point.
Basics
I want it to be highly abstract - I don’t care if it’s a forest, if it’s Cinderella etc., I want the relationships.
Let’s try. Cinderella is basically “Rags to riches”, so:
- Character roles
- Protagonist
- Underprivileged protagonist (Cinderella)
- Benefactor
- …
- Protagonist
- Key plot points
- Hardship
- Opportunity
- Transformation
- Achievement
- Thematic elements
- Transformation
- Reward?..
…
Or GPT3’s ideas from before:
"Entities": { "Thief": {"Characteristics": ["Cunning", "Resourceful"], "Role": "Protagonist"}, "Fish": {"Characteristics": ["Valuable", "Symbolic"], "Role": "Object"}, "Owner": {"Characteristics": ["Victimized", "Unaware"], "Role": "Antagonist"} }, "Goals": { "Thief": "Steal Fish", "Owner": "Protect Property" }, "Challenges": { "Thief": "Avoid Detection", "Owner": "Secure Property" }, "Interactions": { ("Thief", "Fish"): "Theft", ("Thief", "Owner"): "Avoidance", ("Owner", "Fish"): "Ownership" }, "Outcomes": { "Immediate": "Successful Theft", "Long-term": "Loss of Trust" }, "Moral Lessons": { "Actions Have Consequences", "Importance of Trust", "Greed Leads to Loss" }- ENTITIES have a:
- ROLE
- Protagonist
- CHARACTERISTICS
- Poor
- ROLE
Awesome chats with ChatGPT
Here’s it generating an ontology based on the above graph: https://chat.openai.com/share/92ed18ce-88f9-4262-9dd9-f06a07d06acc
And more in UKR: https://chat.openai.com/share/846a5e85-353e-4bb5-adbe-6da7825c51ed
Tale ontology v0.0.1
In bold bits I’m not sure of. In decreasing order of abstraction, with the first two being the most generic ones and the latter ones more fitting for concrete stories.
- KEY PLOT POINTS:
- Hardship
- Disruption
- Opportunity
- Transformation
- Achievement
- CHARACTER ROLES:
- Description: most abstract roles for the characters
- Values:
- Protagonist
- Antagonist
- Object
- Benefactor
- Goal object
- Ally
- etc.
- ENTITIES
- Description:
- can be characters, important objects, whatever
- Attributes:
Characteristics:- … basically adjectives.
- e.g. Resourceful, motivated, hungry
Role: CHARACTER ROLE
- Description:
- GOALS
- Description: what drives the character/entity
- Attributes:
Entity: ENTITYGoal: main goal of entity in this context
- Example:
- Thief: steal fish
- Owner: protect property
- INTERACTIONS
- Plaintext description of what’s up between pairs of ENTITIES
- Example: (’thief’,‘fish’): theft
- OUTCOMES
- Attributes:
SHORT-TERM: plaintext descriptionLONG-TERM: plaintext description
- Example: “loss of trust”
- Optional
- Attributes:
- MORAL LESSONS:
- Description: short plaintext description
- Optional
- examples:
- “Actions Have Consequences”, “Importance of Trust”, “Greed Leads to Loss”
Remaining issues:
- Can I generalize to remove “fish” and stuff? Maybe call it object?
- Two versions of the ontology with different levels of abstractions?
Test 1:
Here’s ChatGPT applying that to Shrek: https://chat.openai.com/share/d96d4be6-d42f-4096-a18f-03f786b802c6
Modifying its answers:
“Using this ontology for abstract fairy tale description, please create a generalized graph structure for THE FIRST HARRY POTTER MOVIE. Focus on the overarching themes and character roles without specific names or unique settings. The graph should include key plot points, character roles, entities, goals, interactions, outcomes, and moral lessons, all described in a manner that is broadly applicable to similar stories.”
Revised Ontology for Abstract Fairy Tale Description
Character Roles
- Protagonist: Central character facing a challenge.
- Antagonist: Character or force opposing the protagonist.
- Goal Object: The objective or person the protagonist seeks.
- Ally: A supportive character aiding the protagonist.
Entities
- Description: Characters or significant elements in the story.
- Attributes:
- Characteristics: Abstract adjectives describing the entity.
- Role: One of the defined character roles.
Goals
- Description: Motivations driving the entities.
- Attributes:
- Entity: Reference to an entity.
- Objective: Abstract goal of the entity.
Interactions
- Description: General interactions between pairs of entities.
- Format: (‘Entity1’, ‘Entity2’): Type of interaction.
Outcomes
- Short-term: Immediate result of the story’s climax.
- Long-term: Lasting changes or lessons from the story.
Moral Lessons
- Description: Universal themes or morals derived from the story.
Attempt 2
<
Tools
- Character roles
-
Day 1822 (27 Dec 2023)
Converting markdown thesis to pdf and stuff
Context:
- 230928-1745 Masterarbeit draft,
- 231225-2240 Glosses markdown magic, 231226-1702 Ideas for annotating glosses in my Masterarbeit
> pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.pdf # unicode magic Try running pandoc with --pdf-engine=xelatex. # thank you > pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.pdf --pdf-engine=xelatex # a volley of... [WARNING] Missing character: There is no о (U+043E) in font [lmroman10-italic]:mapping=tex-text;!- Ugly:
-
Makefile magic etc
Exporting Hugo to PDF | akos.ma looks nice.
build/pdf/%.pdf: content/posts/%/index.md $(PANDOC) --write=pdf --pdf-engine=xelatex \ --variable=papersize:a4 --variable=links-as-notes \ --variable=mainfont:DejaVuSans \ --variable=monofont:DejaVuSansMono \ --resource-path=$$(dirname $<) --out=$@ $< 2> /dev/nullLet’s try:
pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.pdf --pdf-engine=xelatex --variable=links-as-notes \ --variable=mainfont:DejaVuSans \ --variable=monofont:DejaVuSansMonoBetter but not much; HTML is not parsed, lists count as lists only after a newline it seems.



pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.pdf --pdf-engine=xelatex --variable=links-as-notes \ --variable=mainfont:DejaVuSans \ --variable=monofont:DejaVuSansMono \ --from=markdown+lists_without_preceding_blanklineBetter, but quotes unsolved:

Markdown blockquote shouldn’t require a leading blank line · Issue #7069 · jgm/pandoc
pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.pdf --pdf-engine=xelatex --variable=links-as-notes \ --variable=mainfont:DejaVuSans \ --variable=monofont:DejaVuSansMono \ --from=markdown+lists_without_preceding_blankline #+blank_before_blockquoteACTUALLY,
- f gfm(github-flavour) solves basically everything.commonmarkdoesn’t parse latex,commonmark_x(‘with many md extensions’) on first sight is similar togfm.I think HTML is the last one.
Raw HTML says it’s only for strict:
--from=markdown_strict+markdown_in_html_blocksmsword - Pandoc / Latex / Markdown - TeX - LaTeX Stack Exchange suggest md to tex and tex to pdf, interesting approach.
6.11 Write raw LaTeX code | R Markdown Cookbook says complex latex code may be too complex for markdown.
This means this except w/o backslashes:
\```{=latex} $\underset{\text{NOUN-NOM}}{\overset{\text{man}}{\text{чоловік-}\varnothing}}$ $\underset{\text{PST}}{\overset{\text{saw}}{\text{побачив}}}$ $\underset{\text{NOUN-ACC}}{\overset{\text{dog}}{\text{собак-у}}}$. \```Then commonmark_x can handle that.
EDIT:
--standalone!More on HTML sub/sup to PDF
I don’t need HTML, I need
<sub>.-
pandoc md has a syntax for this: Pandoc - Pandoc User’s Guide
- …but I’m not using pandoc md :(
-
Options
- Can I replace my tags w/ that with yet another filter?
- Ignore that obsidian/hugo can’t parse them and use pandoc syntax?.. and do
--from=markdown+lists_without_preceding_blankline+blank_before_blockquote? :( - export to HTML w/ mathjax and from it PDF?
- just use latex syntax everywhere? :(
ChatGPT tried to create a filter but nothing works, I’ll leave it for later: https://chat.openai.com/share/c94fffbe-1e90-4bc0-9e97-6027eeab281a
HTML
This produces the best HTML documents:
> pandoc 230928-1745\ Masterarbeit\ draft.md -o master_thesis.html \ --from=gfm --mathjax --standaloneNB If I add CSS, it should be an absolute path:
Later
- 6.2 Pandoc options for LaTeX output | R Markdown Cookbook
- This has config options for latex, incl. fonts Pandoc - Pandoc User’s Guide
- 6.4 Include additional LaTeX packages | R Markdown Cookbook what a cool place
Callouts
It’d be cool to wrap examples in the same environment!
https://forum.obsidian.md/t/rendering-callouts-similarly-in-pandoc/40020:
-- https://forum.obsidian.md/t/rendering-callouts-similarly-in-pandoc/40020/6 -- local stringify = (require "pandoc.utils").stringify function BlockQuote (el) start = el.content[1] if (start.t == "Para" and start.content[1].t == "Str" and start.content[1].text:match("^%[!%w+%][-+]?$")) then _, _, ctype = start.content[1].text:find("%[!(%w+)%]") el.content:remove(1) start.content:remove(1) div = pandoc.Div(el.content, {class = "callout"}) div.attributes["data-callout"] = ctype:lower() div.attributes["title"] = stringify(start.content):gsub("^ ", "") return div else return el end endMakes:
> [!NOTE]- callout Title > > callout contentinto
::: {.callout data-callout="note" title="callout Title"} callout content :::.callout { color: red; /* Set text color to red */ border: 1px solid red; /* Optional: add a red border */ padding: 10px; /* Optional: add some padding */ /* Add any other styling as needed */ }Then this makes it pretty HTML:
pandoc callout.md -L luas/obsidian-callouts.lua -t markdown -s | pandoc --standalone -o some_test.html --css luas/callout-style.css<div class="callout" data-callout="note" title="callout Title"> <p>callout content</p> </div>For PDF: .. it’s more complex, will need such a header file etc. later on. TODO
\usepackage{xcolor} % Required for color definition \newenvironment{callout}{ \color{red} % Sets the text color to red within the environment % Add any other formatting commands here }{}Unrelated
Footnotes
- What if I put the footnotes in the text margins and do a HTML-first thesis?
- margin notes!
Tufte CSS with pandoc
- A Tufte Handout Example
- tufte-css/tufte.css at gh-pages · edwardtufte/tufte-css
- Which I can add as
--css /abs/tufte.css!
- Which I can add as
- even better: jez/tufte-pandoc-css: Starter files for using Pandoc Markdown with Tufte CSS
- hard install process though
- very hard
- It uses this: jez/pandoc-sidenote: Convert Pandoc Markdown-style footnotes into sidenotes
Copied executables to /home/sh/.local/bin/:aha so that’s where you put your filters, inside$PATH
Damn! Just had to replace index.md with my thesis, then
make alland it just …worked. Wow.
Apparently to make it not a sidenote I just have to add
-to the footnote itself. Would be trivial to replace with an@etc., then I get my inital plan - citations as citations and footnotes with my remarks as sidenotes.I can add
--from gfm --mathjaxto the makefile command and it works with all my other requirements!pandoc \ --katex \ --section-divs \ --from gfm \ --mathjax \ --filter pandoc-sidenote \ --to html5+smart \ --template=tufte \ --css tufte.css --css pandoc.css --css pandoc-solarized.css --css tufte-extra.css \ --output docs/tufte-md/index.html \ docs/tufte-md/index.mdI wonder if I can modify it to create latex-style sidenotes, it should be very easy: pandoc-sidenote/src/Text/Pandoc/SideNote.hs at master · jez/pandoc-sidenote
Numering references etc
{#fig:label} $$ math $$ {#eq:label} Section {#sec:section}TODO figure out, and latex as well.
Citations
TODO
-
Day 1821 (26 Dec 2023)
Ideas for annotating glosses in my Masterarbeit
Current best:
eng: the manNOM.SG saw the dogNOM.SG
ukr: чоловікman-NOM.SG побачивsaw-PST собакydog-ACC.SGIdeas
I’d love to integrate the usual UD feats bits but they take a lot of space, and it’s either latex magic or one word per line.
-
ukr: чоловік(man): Case=Nom|Number=Sing побачив(saw) собакy(dog): Case=Acc|Number=Sing
-
$чоловік^{man}_{Case=Nom|Number=Sing}$
-
${\underset{man}{чоловік}}^{Case=Nom|Number=Sing}$
-
$\underset{Case=Nom|Number=Sing}{чоловік^{man}}$
-
$\underset{NOM.SG}{чоловік^{man}}$
-
${\underset{man}{чоловік}}^{Case=Nom|Number=Sing}$
-
${\underset{man}{чоловік}}^{NOM.SG}$
-
${\underset{man}{чоловік}}^{NOM.SG}$ ${\underset{saw}{побачив}}$ ${\underset{dog}{собаку}}^{GEN.PL}$
я I Case=Nom|Number=Sing
побачив saw
собаку saw Animacy=Anim|Case=Acc|Gender=Masc|Number=Singukr: чоловікman-NOM.SG побачивsaw-PST собакydog-GEN.PL
${\underset{man}{чоловік}}$ Case=Nom|Number=Sing ${\underset{man}{чоловік}}$ Case=Nom|Number=Sing
I think this is cool! But hell to write and parse:
$\underset{\text{NOUN.NOM}}{\overset{\text{man}}{\text{чоловік-}\varnothing}}$ $\underset{\text{PST}}{\overset{\text{saw}}{\text{побачив}}}$ $\underset{\text{NOUN-ACC}}{\overset{\text{dog}}{\text{собак-у}}}$.
$\underset{\text{NOUN.NOM}}{\overset{\text{man}}{\text{чоловік-}\varnothing}}$ $\underset{\text{PST}}{\overset{\text{saw}}{\text{побачив}}}$ $\underset{\text{NOUN-ACC}}{\overset{\text{dog}}{\text{собак-у}}}$.Let’s play more with it:
$\underset{\text{Case=Nom|Number=Sing}}{\overset{\text{man }}{\text{чоловік}}}$ $\underset{\text{}}{\overset{\text{saw}}{\text{побачив}}}$ $\underset{\text{Case=Acc|Number=Sing}}{\overset{\text{dog}}{\text{собаку}}}$.
I can split it in diff lines: $\underset{\text{Case=Nom|Number=Sing}}{\overset{\text{man }}{\text{чоловік}}} \underset{\text{}}{\overset{\text{saw}}{\text{побачив}}} \underset{\text{Case=Acc|Number=Sing}}{\overset{\text{dog}}{\text{собаку}}}$.
$$\underset{\text{Case=Nom|Number=Sing}}{\overset{\text{man }}{\text{ЧОЛОВІК}}} \underset{\text{}}{\overset{\text{saw}}{\text{ПОБАЧИВ}}} \underset{\text{Case=Acc|Number=Sing}}{\overset{\text{dog}}{\text{СОБАКУ}}}$$
Splitting by morphemes
ukr: використовуватимуться Aspect=Imp|Number=Plur|Person=3
1 використовуватимуться використовуватися VERB _ Aspect=Imp|Mood=Ind|Number=Plur|Person=3|Tense=Fut|VerbForm=Fin 0 root _ SpaceAfter=Noukr: використовуватимуть-сяVERB-REFL
ukr: використовуватимутьVERB -сяREFL
$\underset{\text{NOM.SG}}{\overset{\text{man }}{\text{чоловік}}}$ $\underset{\text{PST}}{\overset{\text{saw}}{\text{побачив}}}$ $\underset{\text{SG-ACC}}{\overset{\text{dog}}{\text{собак-у}}}$.
Text representation of graphs with graphviz
This is one of the cooler ones, I’ll use it if I ever need to: Examples — graphviz 0.20.1 documentation
It’s also supported by HackMD! How to use MathJax & UML - HackMD
Markdown collaboration tools
I’ll need something like overleaf for my markdown thesis.
5 Best Collaborative Online Markdown Editors - TechWiser
- HackMD
- Markdown quick start guide - HackMD is nice!
- The comments are part of the markdown code itself
- but no way to highlight a certain phrase, just add a comment in a place of the text
- you can share your doc to make it editable to not-logged-in users
- Seems to hold my current thesis length quite well
- I think I can stop looking
-
-
Day 1820 (25 Dec 2023)
Glosses markdown magic
Interlinear Glosses
… are a way to annotate grammar bits of a language together with translation: Interlinear gloss - Wikipedia
The Leipzig Glossing Rules are a set of rules to standardize interlinear glosses. They are focused less on understandability and more on consistency.
- Markdown
- cysouw/pandoc-ling: Pandoc Lua filter for linguistic examples
- gunnarnl/pangb4e: Pandoc filter for gb4e support
- parryc/doctor_leipzig: Leipzig, MD - glossing for Markdown
- one can always do
<span style="font-variant:small-caps;">Hello World</span>[^1]
- Python
Using interlinear glosses
I’m writing my thesis in Obsidian/Markdown, synced to Hugo, later I’ll use sth like pandoc to make it into a PDF, with or without a latex intermediate step.
EDIT: newer technical part lives now here 231226-1702 Ideas for annotating glosses in my Masterarbeit
Pandoc-ling
cysouw/pandoc-ling: Pandoc Lua filter for linguistic examples
> pandoc --lua-filter=pandoc_ling.lua 231225-2240\ Glosses\ markdown\ magic.pandoc.md -o test.pdf Error running filter pandoc_ling.lua: pandoc_ling.lua:21: attempt to call a nil value (method 'must_be_at_least') stack traceback: pandoc_ling.lua:21: in main chunk:::ex | Dutch (Germanic) | Deze zin is in het nederlands. | DEM sentence AUX in DET dutch. | This sentence is dutch. ::::::ex | Dutch (Germanic) | Deze zin is in het nederlands. | DEM sentence AUX in DET dutch. | This sentence is dutch. :::
.. it was the pandoc version. Updated. No error, but no luck either.
Digging into the examples I think this is happening:
Code is code. Using that formatting without code makes it be interpreted as a line, and that doesn’t survive the obsidian’s pandoc extensions’ conversion to pandoc markdown.
The original docu generation had this script:
function addRealCopy (code) return { code, pandoc.RawBlock("markdown", code.text) } end return { { CodeBlock = addRealCopy } }It changes code blocks into code blocks and the content of the code block. Then the
:::block is put after the code but like normal markdown text, and it gets correctly changed by the pandoc-ling filter.> pandoc 231225-2240\ Glosses\ markdown\ magic.pandoc.md -t markdown -L processVerbatim.lua -s --wrap=preserve | pandoc -L pandoc_ling.lua -o my.htmlI can drop exporting extensions and just manually convert bits?..
This works:
> pandoc "garden/it/231225-2240 Glosses markdown magic.md" -t markdown -L pandoc_ling.lua -s > pandoc "garden/it/231225-2240 Glosses markdown magic.md" -L pandoc_ling.lua -o my.htmland is OK if my masterarbeit file will have no complexities at all.
(Can i add this as parameter to the existing bits?)
YES!
-L /home/sh/t/pandoc/pandoc_ling.luaadded as option to the pandoc plugins, together with “from markdown” (not HTML) option, works for getting this parsed right!
(Except that it’s ugly in the HTML view but I can live with that)
And Hugo. Exporting to Hugo through obyde is ugly as well.
I colud write sth like this: A Pandoc Lua filter to convert Callout Blocks to Hugo admonitions (shortcode).
We’lll o
Obsidian link gloss plugin
Mijyuoon/obsidian-ling-gloss: An Obsidian plugin for interlinear glosses used in linguistics texts.
Pandoc export from HTML visualizes them quite well.
\gla Péter-nek van egy macská-ja \glb pe:tɛrnɛk vɒn ɛɟ mɒt͡ʃka:jɒ \glc Peter-DAT exist INDEF cat-POSS.3SG \ft Peter has a cat.\gla Péter-nek van egy macská-ja \glb pe:tɛrnɛk vɒn ɛɟ mɒt͡ʃka:jɒ \glc Peter-DAT exist INDEF cat-POSS.3SG \ft Peter has a cat.\set glastyle cjk \ex 牆上掛著一幅畫 / 墙上挂着一幅画 \gl 牆 [墙] [qiáng] [wall] [^[TOP] 上 [上] [shàng] [on] [^]] 掛 [挂] [guà] [hang] [V] 著 [着] [zhe] [CONT] [ASP] 一 [一] [yì] [one] [^[S] 幅 [幅] [fú] [picture.CL] [] 畫 [画] [huà] [picture] [^]] \ft A picture is hanging on the wall.Maybe a solution
- Do all my glosses using pandoc-ling format
- put them into code blocks belonging to a special class
- write a very similar filter to processVerbatim but that operates only on code blocks of this class
- when outputting to Hugo they’ll stay as preformatted code
- when exporting to pandoc run them through it first, then pandoc-ling, leading to pretty glosses in the final exported option
function addRealCopy (code) -- return { code, pandoc.RawBlock("markdown", code.text) } if code.classes[1] == "mygloss" then return { pandoc.RawBlock("markdown", code.text) } else return { code } end end return { { CodeBlock = addRealCopy } }Should parse:
:::ex | Dutch (Germanic) | Deze zin is in het nederlands. | DEM sentence AUX in DET dutch. | This sentence is dutch. :::Should stay as code:
:::ex | Dutch (Germanic) | Deze zin is in het nederlands. | DEM sentence AUX in DET dutch. | This sentence is dutch. :::pandoc "/... arden/it/231225-2240 Glosses markdown magic.md" -L processVerbatim.lua -t markdown -s | pandoc -L pandoc_ling.lua -o my.htmlIt works!
But not this:
> pandoc "/home231225-2240 Glosses markdown magic.md" -L processVerbatim.lua -L pandoc_ling.lua -o my.htmlLikely because both require markdown and the intermediate step seems to break.
Maybe I’m overcomplicating it and I can just use the UD I can use superscripts!
Just use superscripts
The inflectional paradigm of Ukrainian admits free word order: in English the Subject-Verb-Object word order in “the manman-NOM.SG saw the dogdog-NOM.SG” (vs “the dog man-NOM.SG saw the manman-NOM.SG “) determines who saw whom, while in Ukrainian (“чоловікman-NOM.SG побачивsaw-PST собакУdog-GEN.PL”) the last letter of the object (dog) makes it genetive, and therefore the object.
- Markdown