serhii.net
In the middle of the desert you can say anything you want
-
Day 1816 (21 Dec 2023)
Perceptual image hashes
Related: 231220-1232 GBIF iNaturalist plantNet duplicates
KilianB/JImageHash: Perceptual image hashing library used to match similar images does hashes based on image content, not bytes (a la SHA1 and friends)
Hashing Algorithms · KilianB/JImageHash Wiki is a cool visual explanation of the algos involved.
Kind of Like That - The Hacker Factor Blog is a benchmark thing, TL;DR
- aHash is very quick but many FP
- dHash just as quick but better
One of the comments suggest running a quick one with many FPs and then a slower one on the problematic detected images.
-
Day 1815 (20 Dec 2023)
GBIF iNaturalist plantNet duplicates
TL;DR
- plantNet does no deduplication I could find
- iNat
- allows literally duplicating observations with a click e.g. if there are multiple plants to detect, keeping image files constant
- etiquette allows duplicating observations in many cases, unless it’s e.g. literally the same photo (but diff observations for different growth stages is fine)
- good karma to mention related observations in the description
- does no deduplication by images etc. on its side, but may soon have something
- GBIF
- is more worried about adding the same herbarium plant twice from different collections or same picture both from iNat and Pl@ntNet
- does deduplication more like record linking
- has a clustering/collection feature but it looks for similar observations only across datasets, not within the same one. But the blog post about it is awesome and usable to do deduplication within same datasets as well.
- is more worried about adding the same herbarium plant twice from different collections or same picture both from iNat and Pl@ntNet
- Recommendation: roll SHA1 ourselves
iNaturalist
Fazit
- duplicates can happen if:
- bad: multiple pics of the same organism at same time as different observations
- good: different organisms on the same pic
- using the duplicate function -> URI seems to be preserved
- manually upload picture, possibly cropped
- etiquette seems to be that if it’s different parts of picture add it to description
- we can look for links to observations in descriptions
- no auto-duplication detection included in iNat. Wikipedia uses SHA1 for this purpose.
Refs
-
Finally, you can totally upload the same picture multiple times if there’s multiple organisms in one picture that you would like identified - you want to have a separate observation for each organism. Usually if I do this I’ll make a note in the description of what I’m looking to have IDed.
- duplicate observations OK if same plant different time etc.
-
https://www.inaturalist.org/posts/28325-tech-tip-tuesday-duplicating-observations: “perfectly okay to duplicate observations”, because species in the background etc.
- there’s literally a function for this
- otherwise one can crop the picture
-
https://forum.inaturalist.org/t/duplicate-obervations/18378/4
- wikimedia uses SHA1 to identify duplicates
-
- inat doesn’t have duplicate detection for pictures uploaded multiple times (discussion on adding this feature)
-
Random
-
just came across a user who repeatedly submits pairs of some robber fly photo weeks apart. (https://forum.inaturalist.org/t/create-a-flag-category-for-duplicate-observations/29647/42)
-
Example instances
- same pic, different crops:
- Other observations mentioned in descriptions
PlantNet
- TL;DR nothing.
- Found nothing useful after a quick google search
- Pl@ntNet automatically identified occurrences
- TODO understand what is that, but I think it’s about requests than about plants:
remove shared queries (already present in observation dataset) - remove duplicate session (keep the most recent query based on the session number) -
- TODO understand what is that, but I think it’s about requests than about plants:
GBIF
Fazit
- GBIF has clustering of records to find duplicates, but does it only between datasets, not within the same one
- REALLY cool pages on the topic and in general: Identifying potentially related records - How does the GBIF data-clustering feature work? - GBIF Data Blog
- Examples
- how to search for it: this is all plantnet observations that are part of a cluster Occurrence search (“has related records”)
- observation that is part of a cluster Occurrence 4011692074
- Darwin Core has
associatedOccurrencesfor kinda similar ones, incl. by parsing the descriptions for mentions etc.: Darwin Core Quick Reference Guide - Darwin Core- not used for duplicate detection as of the awesome blog post above from 2021-11-04
- some people use it in other ways: https://discourse.gbif.org/t/differences-in-use-between-associatedoccurrences-and-associatedorganisms/2561/2
- I can’t find how to access it from the API
- Their main threat model is the same herbarium speciment scanned and put into different collections, or the same plant photo uploaded to two different platforms (e.g. pNet + iNat) ending in GBIF twice
Refs
-
Has clustering of records that appear to be similar:
- Identifying potentially related records - How does the GBIF data-clustering feature work? - GBIF Data Blog
- very thorough
- downsides
- clustering compares only across datasets, not within the same one!
- runs once every few weeks
- Biodiversity Data Use (less cool page mentioning duplications)
- More theory on this: New data-clustering feature aims to improve data quality and reveal cross-dataset connections
- Occurrence search (“has related records”)
- Example: Occurrence 4036191318
- in API:
- New data-clustering feature aims to improve data quality and reveal cross-dataset connections
matching similar entries in individual fields across different datasets
- https://api.gbif.org/v1/occurrence/1141981356/experimental/related
- vanilla
curl https://api.gbif.org/v1/occurrence/4011664186 | jq -C | lesshasisInClusterbut nothing more
- New data-clustering feature aims to improve data quality and reveal cross-dataset connections
- Identifying potentially related records - How does the GBIF data-clustering feature work? - GBIF Data Blog
-
GBIF has
associatedOccurrences- Darwin Core Quick Reference Guide - Darwin Core
- not yet used in clustering
- parses descriptions etc. for links
-
Darwin Core Resource Relationship – Extension darwin core bit about relationships between records docu
-
Duplicate occurrence records - Data Publishing - GBIF community forum
- user distinguishes three types of duplicates: exact, strict, relaxed
- discussions of using bash/cli magic to parse occurrences.txt to find them,
- ref. A data cleaner’s cookbook - Content 1
- BASHing data: Partial duplicates
- all with a more record linking vibe
-
I’m not aware of any backend or external packages (e.g. in R or Python) that can tidy a Darwin Core dataset
-
Fortunately or unfortunately, Darwin Core datasets are complex beasts that don’t lend themselves to automated checking and fixing. For this reason people (not backend routines) are the best Darwin Core data cleaners 4. The code recipes 2 I use are freely available on the Web and I (and now others) are happy to train others in their use.
-
Duplicate observations across datasets - GBIF community forum
-
Something that I often hear repeated on iNaturalist and BugGuide is that posting the same observation on both platforms results in the observation being ingested twice by GBIF.
- TL;DR posted twice, not found as duplicate because had no location info on one of the platforms
-
Random
- Developer Blog: “I noticed that the GBIF data portal has fewer records than it used to – what happened?” about removing dulicates from GBIF in 2012
GBIF/iNat
Looking for observations with links in description
- creeping thistle from Norfolk County, ON, Canada on July 12, 2019 at 08:44 AM by Jeremy Hussell. Same plants as in this previous observation: https://www.inaturalist.org/observations/28646599 · iNaturalist == Occurrence 3005247358
- “occurrence remarks” in GBIF are iNat’s “description”
Random / fun
- https://community.openstreetmap.org/t/semi-automated-tree-additions/102197/57?page=4 openstreetmap automatically adding trees and needing deduplication
Next steps
Look for GBIF/iNat/plantnet repos on Github and look their mentions of duplicates
-
Day 1812 (17 Dec 2023)
Obsidian has an Outline core plugin
Core plugins -> Outline!
Poetry add spacy model to requirements
Usually models are added as
python -m spacy download de_core_news_smFor poetryi: python - How to download spaCy models in a Poetry managed environment - Stack Overflow
TL;DR: spacy models are python packages!
Get direct link to model packages here: uk · Releases · explosion/spacy-models
Add to poetry tool dependencies in pyproject.toml:
[tool.poetry.dependencies] python = "^3.10" # ... uk_core_news_sm = {url = "https://github.com/explosion/spacy-models/releases/download/uk_core_news_sm-3.7.0/uk_core_news_sm-3.7.0-py3-none-any.whl"}add through poetry CLI:
poetry add https://github.com/explosion/spacy-models/releases/download/uk_core_news_sm-3.7.0/uk_core_news_sm-3.7.0-py3-none-any.whl
-
Day 1810 (15 Dec 2023)
Hierarchical tree list of running processes in linux
I’d usually do
ps aux.The
pscommand can also do:ps -ef --forest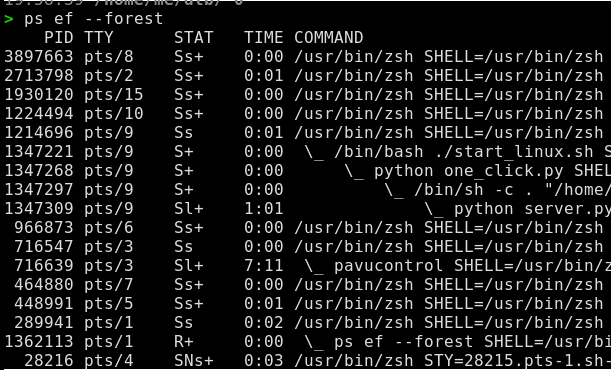
But the best one is
pstree1 from thepsmiscpackage.pstree # or pstree -i # for processids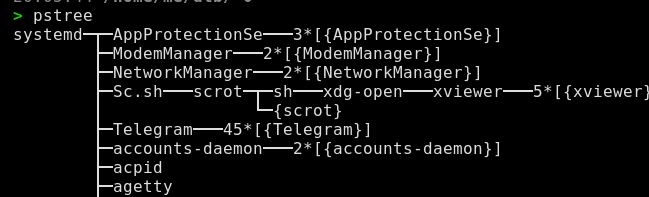
-
Day 1809 (14 Dec 2023)
Asking ChatGPT to make its own prompts is a superpower
Used it 3 times already and it’s awesome.
— If I’ll want your help with this in the future, which prompt can I use to describe the task I need and the output type to get a graph in the format and complexity level of the one you just generated? How can I concisely describe it to you so that no clarifications will be needed and you can just give me the answer?
— “Create an abstract graph structure for a story involving multiple characters with interconnected goals, challenges, outcomes, and a moral lesson. The graph should use nodes and relationships similar to the format of the ‘Adventurer and Guide’ mountain climbing story you previously created, with entities, goals, challenges, interactions, outcomes, and a moral lesson. The structure should reflect underlying themes rather than the literal narrative, similar to the complexity and abstraction level of the previous example.”
After more clarifications:
“Generate an abstract graph structure for a narrative involving multiple animate characters. The graph should include nodes for entities, goals, challenges, interactions, outcomes, and moral lessons. Each node should abstractly represent the core elements of the story, focusing on thematic and moral aspects rather than the literal narrative. The format should be similar to a semantic web ontology, emphasizing relationships and abstract concepts. Please provide the graph in a Python dictionary format, with complexity and depth akin to an advanced semantic network.”
Context: 231024-1704 Master thesis task CBT
Masterarbeit benchmark task for Russian-Ukrainian interference
-
231213-1710 Ukrainska Pravda dataset#Can I also use this to generate tasks for the UA-CBT ( 231024-1704 Master thesis task CBT ) task? : both 3.5 and 4 during summarization use definitely Russian-inspired phrases :
-
In the news summarization bit, it magically changed Євген->Евген (https://chat.openai.com/share/2f6cf1f3-caf5-4e55-9c1b-3dbd6b73ba29)
-
Та подивись, баране, як я виглядаю з цим стильним сурдутом1
Вертить хвостиком і крутить рогами. Цап робить враження2.
Old links
(from 230928-1630 Ideas for Ukrainian LM eval tasks)
- СЛОВОВЖИВАННЯ | Горох — українські словники
- відноситися - Антисуржик. Словник «українського» суржика
- Словник-антисуржик онлайн
- Антисуржик (словник) - Русский/украинский язык, культура - Форум Днепродзержинск-Каменское
EXCELLENT! Мова – не калька: словник української мови - Тарас Береза - Тека авторів - Чтиво- parse -> estimate frequency -> include only the most frequent?
- A lot of the examples there are let’s say questionable to my central-Ukrainian ear
- голий -> “У костюмі (в одежі) Адама і Єви; у чому мати [на світ] народила.” alrighty then
- Льотчик -> летун
- Ліберія -> “Вільна країна” I’m done
- I want RU interference (!= суржик); I want RU interference (!= стилістика)
- Some kind of filtering is definitely needed. Could be as easy as putting “1” in rows of a spreadsheet
- https://chtyvo.org.ua/authors/Tykhyi_Oleksii/Slovnyk_movnykh_pokruchiv.pdf
- Суржиково-український словник
- has really nice intro!
- Українське життя в Севастополi Юрій Гнаткевич СЛОВНИК-АНТИСУРЖИК ^ff5ccc
- Frame as multiple-choice task! Or boolean? Or “Is this a correct sentence”?
- I really like this: `“Цей студент [взявся за/почав] дослідження важкої теми.”
- For fun, here’s ChatGPT lying about prefixes: https://chat.openai.com/share/0eda9061-d2cf-46bc-ad45-38cc6e58934a
- False friends!
- Here’s an itemized list: Фальшиві друзі перекладача — Вікіпедія
- сир/сыр, неділя/неделя/…
- False Friends of the Slavist/Russian-Ukrainian - Wikibooks, open books for an open world
- ChatGPT ideas:
-
On the semantic front, exploit polysemy and homonymy differences. Formulate sentences with words that have multiple meanings in Russian, but those meanings have distinct equivalents in Ukrainian. This will challenge the model to accurately discern the intended sense based on context.
More ideas
Using correct English spelling of cities etc!
-
-
Day 1808 (13 Dec 2023)
Ukrainska Pravda dataset
This post describes the Ukrainska Pravda dataset I created as part of my Master’s Thesis. The contents of this blog post will be edited (esp. for brevity) and become part of the thesis (230928-1745 Masterarbeit draft).
Ukrainska Pravda articles classification
A novel dataset created in the context of this Master’s Thesis is the Ukrainska Pravda multilingual dataset. The package written for this,
UPCrawler, is released at (https://github.com/pchr8/up_crawler) under the MIT license.The dataset is released on the HF Hub at https://huggingface.co/datasets/shamotskyi/ukr_pravda_2y / doi https://doi.org/10.57967/hf/1476 under the CC BY-NC 4.0 license.
Ukrainska Pravda
Ukrainska Pravda (lit. “Ukrainian Truth”; https://www.pravda.com.ua/) is a Ukrainian online newspaper for a general readership writing, mostly, about political and social topics.
In 2017, it was in the eighth most cited source of the Ukrainian Wikipedia1 and in 2020 it was the most visited online news website in Ukraine2(TODO - better source). The Institute of Mass Information listed Ukrainska Pravda listed it among the six online editions with the highest level of compliance with professional journalistic standards in 2021.3
Website structure
UP (Ukrainska Pravda) publishes articles predominantly in Ukrainian, with some being translated to Russian and English. Each article can belong to zero or more “topics” (tags) that are mostly preserved across translations.
Each article has an article ID that is constant across translations.
Crawling
The crawler interface
The CLI interface expects a date range (using natural language, e.g. “last year”) and a target folder, where the pages are saved.
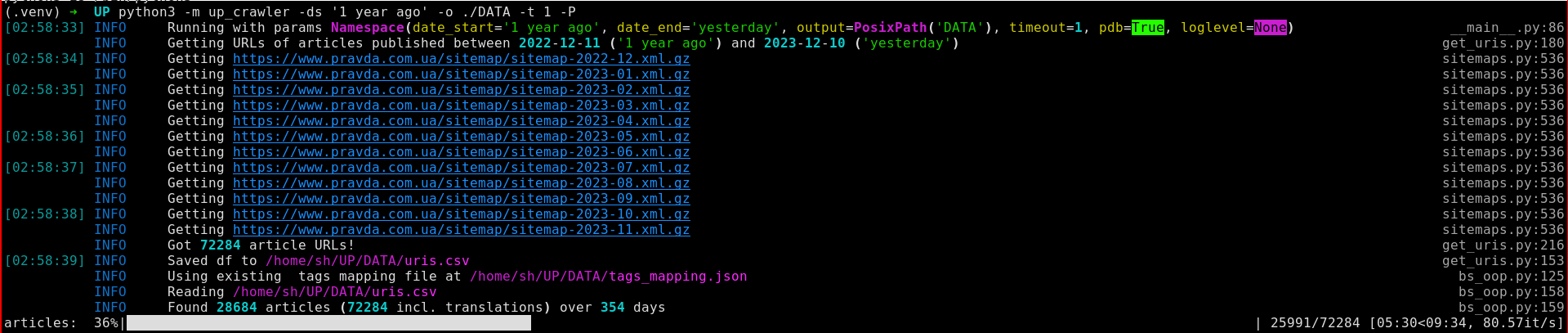
Getting URLs of articles to crawl with Sitemaps
Initially, the package
UPCrawlerused the daily archive pages (e.g. https://www.pravda.com.ua/archives/date_27092023/) to get the URLs of articles published on a specific day, then for each article URL accessed the expected locations of the Russian and English translations to check if a translation exists. Later, I rewrote the code to use a much better solution: parsing the XML sitemaps (e.g. https://www.pravda.com.ua/sitemap/sitemap-2023-09.xml.gz) using the advertools Python package.Sitemaps4 is a XML-based protocol used to inform search engines about the URLs available for web crawling, as well as provide additional information about it such as when was the page last updated, how often does the content change, etc.
The following regex (see https://regex101.com/r/dYlIiF/4 for an interactive analysis) is used to parse each URL to get the language of the article, the article ID, the section (news, podcasts, ..) etc.:
URI_REGEX_STR_EXT = r"(?P<uri>(?P<domain>.*\.com\.ua\/)(?P<lang>(eng)|(rus))?\/?(?P<kind>.*?)\/(?P<art_id>.*(?P<date_part>....\/..\/..?)\/(?P<id>.*)\/))"Crawling the individual articles
Crawling the articles is done using the beautifulsoup4 library. I considered the alternative option of using the newspaper3k package which was able to detect the article, title and metadata from UP surprisingly well, but it incorrectly detected some fields (which would have required manual fixes anyway), so I decided to keep my from scratch implementation.
For transparency and in the spirit of ethical crawling5, there were timeouts between requests, and the unique useragent contained a short explanation of my project as well as my email. At no point was I ever contacted or the crawler blocked.
The most challenging part were the tags. The URL of each tag contained a unique identifier that was consistent between translations.
Processing steps
The article text inside
<article>was taken from each page. The content of the tags<p>and<li>were used to extract the plaintext while avoiding advertisements, infoboxes etc.Paragraphs matching some standard article endings like “follow us on Twitter” weren’t added to the plaintext, but not all such endings were filtered out.
The tags required special care because they presented two problems:
- There were pages with lists of tags in Ukrainian and Russian6 but not English
- Some tags had translations to other languages, some didn’t.
Since this was supposed to be a multilingual dataset I wanted to have a list of tags for each article independent on the translations. The solution at the end was to crawl Ukrainian and Russian tags pages to save the short unique ID and both translations, and add English translations to the short IDs when they were seen in the English translations of articles.
An example tag and three translations:
{"ukr":["флот","/tags/flot/"],"rus":["флот","/rus/tags/flot/"],"eng":["naval fleet","/eng/tags/flot/"]}The UPravda multilingual dataset
Dataset description
The UPravda multilingual dataset contains in total XX individual translations of YY articles. X articles have a Ukrainian version, Y a Russian and Z an Engish one.
The dataset has X individual tags, of which the most common ones are shown in the table below: TODO
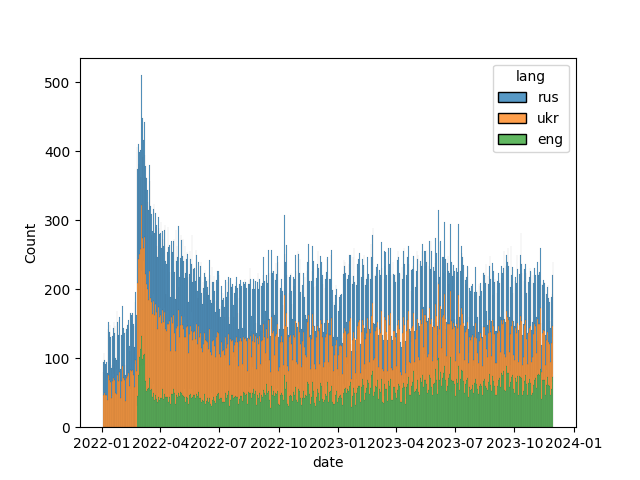
The dataset contains articles published from the 01.01.2022 to X, since UP drastically increased the amount of articles translated to English after the start of the full-scale invasion on the 24.02.2022 7 , (see picture below; TODO better x axis angle on plot).
Mitigations of issues found in multilingual datasets
A recent (2022) manual audit of available crawled multilingual datasets found surprisingly low amounts of in-language data and systematic issues in many of them. 8
Some issues raised in the paper in the context of this dataset:
- Using standard unambiguous ISO 639-3 language codes (ukr, rus, eng). ISO 639-3 was chosen instead of the more common ISO 639-1 (uk, ru, en) because of the possibly ambiguous ‘uk’ that can be associated with Great Britain as well. Interestingly, the more familiar ‘UA’ is a valid ISO code for the country, but not the language.
- The language identification was performed from the URL of the page (in turn labeled by UP), not through automated language identification processes (especially relevant in light of the ukr/rus disambiguation issues discussed in section XXX)
- The texts themselves were written by proficient language users, as opposed to automated translations.
- The dataset is digital-first(TODO word for this): no errors were introduced by OCR, incorrect layout parsing(TODO cite FinDE) or similar.
- I manually checked random articles from the dataset to make sure the different translations are indeed text, in the correct languages, and actually refer to the same article.
Licensing questions
According to Ukrainian law, newpaper-like articles aren’t subject to copyright. According to UP’s rules on the matter9, reprinting (..) in other online-newspapers is free but requires a link to the UP article not later than the second paragraph. Using the materials for commercial reasons is forbidden.
I believe releasing this dataset under the CC BY-NC 4.0 license (that allows sharing and adaptation only with attribution and for non-commercial use), with clear attribution to UP in the name and the description of the dataset, fulfills the applicable obligations both in letter and in spirit.
The dataset is released at https://huggingface.co/datasets/shamotskyi/ukr_pravda_2y
Similar datasets
- UA News classification - ua-datasets / fido-ai/ua-datasets: A collection of datasets for Ukrainian language
- Yehor/ukrainian-news-headlines · Datasets at Hugging Face
- zeusfsx/ukrainian-news · Datasets at Hugging Face
Appendix A: regexes for skipping paragraphs in UPravda dataset
Some UP articles have short paragraphs in the style of “follow us on Twitter” at the end. They have little to do with the actual article, so they were removed from the article text in the dataset.
All paragraphs containing text matching any of the lines/regexes below were filtered out:
"Follow (us|Ukrainska Pravda) on Twitter", "Support UP", "become our patron", "(читайте|слухайте|слушайте) (також|также)", # "read/listen also to", in Russian and Ukrainian
Ways to make a downstream task out of this
- Tags
- News title|text -> tag
- cons: the tags UP uses seem chaotic and inconsistent?…
- Title
- Match title to news text
- Match title to rephrased/summarized news text
ChatGPT prompts for rephrasing the news
It suggested (https://chat.openai.com/share/2f6cf1f3-caf5-4e55-9c1b-3dbd6b73ba29) to me this prompt:
Будь ласка, перефразуйте цей текст, змінюючи порядок інформації та структуру повідомлення, уникаючи збігів слів та фразових конструкцій з оригіналом. Фокусуйтеся лише на ключових фактах, уникаючи зайвих деталей:
An improved version that seems to work ~better(https://chat.openai.com/share/14f12f87-50a8-438c-9d01-a0b076c3be12) :
Будь ласка, перефразуйте цей текст, змінюючи порядок інформації та структуру повідомлення, максимально уникаючи збігів слів та фразових конструкцій з оригіналом. Довжина статті має бути приблизно такою ж, як довжина оригіналу.
GPT3.5 works just as well if not better than GPT4 (and is much faster): https://chat.openai.com/share/78927782-25fa-4047-b2a4-fd01ee9a7a54
Can I also use this to generate tasks for the UA-CBT (231024-1704 Master thesis task CBT) task?
Here GPT4 is much better than GPT3. Can’t share either link because “disabled by moderation”(???).
Interestingly, GPT3.5 used definitely Russian chiches that I document in 231214-1251 Masterarbeit benchmark task for Russian-Ukrainian interference.
Eval downstream task decision
231010-1003 Masterarbeit Tagebuch#2023-12-15
- Solution: article text -> title, out of X options
- give ~10 options with
- ~3 random from the dataset
- ~7 from similar articles from the dataset, e.g. all of the same topic ‘war’
- give ~10 options with
-
<_(@inbook) “Analysis of references across wikipedia languages” (2017) / Włodzimierz Lewoniewski, Krzysztof Węcel, Witold Abramowicz: z / / 10.1007/978-3-319-67642-5_47 _> ↩︎
-
Рейтинг топсайтів України | Інститут масової інформації, linked on Українська правда — Вікіпедія ↩︎
-
Compliance with professional standards in online media. The fourth wave of monitoring in 2021 | Institute of Mass Information ↩︎
-
<_(@Schonfeld2009) “Sitemaps: Above and beyond the crawl of duty” (2009) / Uri Schonfeld, Narayanan Shivakumar: z / https://dl.acm.org/doi/10.1145/1526709.1526842 / 10.1145/1526709.1526842 _> ↩︎
-
Ethics in Web Scraping. We all scrape web data. Well, those of… | by James Densmore | Towards Data Science ↩︎
-
https://www.pravda.com.ua/tags/; https://www.pravda.com.ua/rus/tags/ ↩︎
-
<_(@10.1162/tacl_a_00447) “Quality at a glance: An audit of web-crawled multilingual datasets” (2022) / Julia Kreutzer, Isaac Caswell, Lisa Wang, Ahsan Wahab, Daan van Esch, Nasanbayar Ulzii-Orshikh, Allahsera Tapo, Nishant Subramani, Artem Sokolov, Claytone Sikasote, Monang Setyawan, Supheakmungkol Sarin, Sokhar Samb, Benoît Sagot, Clara Rivera, Annette Rios, Isabel Papadimitriou, Salomey Osei, Pedro Ortiz Suarez, Iroro Orife, Kelechi Ogueji, Andre Niyongabo Rubungo, Toan Q. Nguyen, Mathias Müller, André Müller, Shamsuddeen Hassan Muhammad, Nanda Muhammad, Ayanda Mnyakeni, Jamshidbek Mirzakhalov, Tapiwanashe Matangira, Colin Leong, Nze Lawson, Sneha Kudugunta, Yacine Jernite, Mathias Jenny, Orhan Firat, Bonaventure F. P. Dossou, Sakhile Dlamini, Nisansa de Silva, Sakine Çabuk Ballı, Stella Biderman, Alessia Battisti, Ahmed Baruwa, Ankur Bapna, Pallavi Baljekar, Israel Abebe Azime, Ayodele Awokoya, Duygu Ataman, Orevaoghene Ahia, Oghenefego Ahia, Sweta Agrawal, Mofetoluwa Adeyemi: z / https://doi.org/10.1162/tacl_a_00447 / 10.1162/tacl_a_00447 _> ↩︎
-
Правила використання матеріалів сайтів Інтернет-холдингу ‘‘Українська правда’’ (Оновлено) | Українська правда ↩︎
-
Day 1806 (11 Dec 2023)
poetry running scripts after building python package
Was looking for a way to do this but it’s part of the batteries included: Pluralsight Tech Blog | Python CLI Utilities with Poetry and Typer
If you define run points in the pyproject.toml
[tool.poetry.scripts] up_get_uris = "up_crawler.get_uris:main" up_crawl_uris = "up_crawler.bs_oop:main" up_run = "up_crawler.__main__:main" up_convert = "up_crawler.up_reader:main"Then once you install the package you built with
poetry buildelsewhere, these commands will be registered as cli commands, and then you’ll be able to just runup_run --helpand it’ll work!
Pytest logging output through CLI
I come back to the topic every once in awhile, but this time How To Use Pytest Logging And Print To Console And File (A Comprehensive Guide) | Pytest With Eric gave me the only solution I’ll ever need:
poetry run pytest --log-cli-level=INFOwhich works as-is without any additional packages etc.