serhii.net
In the middle of the desert you can say anything you want
-
Day 1800 (05 Dec 2023)
Notes from paper review
Specific bits
- Number formats! 89.000 / 89000 / 89,000
- Citations
- bad after a
.! \cite{a,b}(->[2,3]) for multiple citations!
- bad after a
- Hyphen vs. Dash–What’s the Difference? | Grammarly
General insighs
- Reviews are awesome and quite useful!
- Awesome and helpful for learning about gaps
Hyphens vs dashes (em-dash, en-dash)
Theory
Moved theory to rl/garden: Hyphens vs dashes vs en-dash em-dash minus etc - serhii.net1
(It seems I can’t do links to the
garden/portion of the website/obyde/obsidian/vault thing, so moved things below there and doing a hard link from here…)Praxis
Updated my (former mirrorboard / pchr8board / …) dvorak xkb layout (220604-0136 Latest iteration of my custom dvorak-UA-RU xkb layout / 230209-0804 Small update to my keyboard layout / pchr8/dvorak_mirrorboard: A Dvorak version of the MirrorBoard) to get an en-dash and em-dash on level5 of the physical/querty keys N and M, so for example
<S-Latch-M>gives me an em-dash/—. I may update the picture later.I hereby promise to use them as much as possible everywhere to remember which ones they are.
(I always hated small dashes in front of two-letter+ entities because it feels wrong, and an en-dash for such cases removes some of the pressure I surprisingly feel when I have to do that, it kinda matches my intuition that a different symbol is needed for longer compound words for clarity reasons.)
This also reminds me that I have quite a few unused Level3 keys on the right side of the keyboard, maybe I can take a second look at it all.
Latex floating figures with wrapfig
First paper I write with ONE column, which required a change to my usual image inclusion process.
Generally I’d do:
\begin{figure}[t] \includegraphics[width=0.4\linewidth]{images/mypic} \caption{My caption} \label{fig:mylabel} \end{figure}Looking into Overleaf documentation about Inserting Images, found out about
wrapfig. Examples from there:\usepackage{wrapfig} % ... \begin{wrapfigure}{L}{0.25\textwidth} \centering \includegraphics[width=0.25\textwidth]{contour} \end{wrapfigure}The magic is:
\begin{wrapfigure}[lineheight]{position}{width} ... \end{wrapfigure}positionfor my purposes islL/rR. Uppercase version allows the figure to float, lowercase means exactly here (a lahinfigure)The first argument
lineheightis how many lines should the picture used. Has to be determined later, but gets rid of the large amount of whitespace that sometimes appears under it.Also — doesn’t like tops/bottoms of pages, section titles, and enums, and creates absolutely ugly results. This really matters.
Includegraphics positions
As a bonus, position options from includegraphics, stolen from Overleaf and converted to table by ChatGPT:
Parameter Position h Place the float here, i.e., approximately at the same point in the source text t Position at the top of the page b Position at the bottom of the page p Put on a special page for floats only ! Override internal parameters LaTeX uses for determining “good” float positions H Places the float at precisely the location in the LATEX code
Overleaf moving or copying projects
Problem: new overleaf project using new template (so no copying the project through interface->copy). The projects have separate folders with multiple files inside, mostly images.
Previously I discovered that you can import files from another overleaf project, without having to download/upload them individually, but I’m still unsure about how linked they are and what happens if they get deleted from the original project.
Today I discovered a better way: download the overleaf project zip, unzip locally, then drag and drop the (multiple!) files to a newly created folder in the new project, all together.
-
Day 1799 (04 Dec 2023)
Masterarbeit evaluation task new UA grammar and feminitives
Something about the recent changes in UA, both the new 2019 orthography and feminitives [^@synchak2023feminine]
New grammar
- ChatGPT doesn’t use the new official grammar!
Feminitives
- What this is NOT:
- not about the СМІ-кліше a la поліціянтка/автоледі!
- Unknown yet: 2. do I want to touch sexism, fairness etc. in the context of this task?
- Relevant-ish
- “ISSN 2409-1154 Науковий вісник Міжнародного гуманітарного університету. Сер.: Філологія. 2019 No 38 том 3” page 119 is nice on this
- Lists
Getting ChatGPT to generate descriptions
-

-
ChatGPT creating his own feminitives! https://chat.openai.com/share/b7e49b4b-9a58-4bf9-80fa-ec11f551b503 has the absolute joy “фахівецька”.
-
Old attempts:
-
I think this format works well enough: https://chat.openai.com/share/9374e47b-f63c-4f97-bfd4-528b11ea0f68
Наведи будь-ласка однозначні короткі дефініції цим професіям чи словам, так, щоб по ним було однозначно очевидно про яку саме професію йде мова. Зміни дефініції так, щоб вони стали фразами, де мова однозначно йде про жінку. Придумай різні варіанти жінок, про яких йде мова, умовно: моя сестра, моя жінка, колишня однокласниця, імена (Марія, Марія Петрівна, Кассандра). Але придумай і свої різноманітніші приклади. Уникай використання самого слова чи поняття у визначеннях. Уникай слів "фахівецька" чи "спеціалістка". Наприклад: Актор: "Моя жінка виконує ролі на сцені чи екрані" Акушерка: "Марія Петрівна допомагає при пологах" Автор: "Я знаю дівчину, яка пише твори та книжки". Будь творчим. Але професія, про яку іде мова, має все рівно бути однозначно зрозумілою. - - - Зроби це для всіх цих слів: абітурієнтка абстракціоністка автоматниця автомобілістка авторка агітаторка агрономка адвокатка анархіст англієць антрополог асистентка астронавт аптекарМожеш зробити з цього json наступного формату? {'job': '', 'description': '' }I asked it to randomly change some of them to be men, so it doesn’t get additional cues. This seems to confuse it and get at least some feminitives to be wrong (“водій”)
Answers
https://chat.openai.com/share/f8a61785-2d2b-4aeb-a24c-45708e40eee3 The format to get it to answer could be:
Я дам список речень, для кожного наведи одне слово, яке це речення логічно завершує. Це має бути іменник, назва ролі або професії. "Моя колишня однокласниця готується до вступу у виш, здібна та наполеглива. Вона - " "Мій брат творить вражаючі образи, захоплюючи глядачів абстракцією. Він - " "Марія Петрівна керує автоматизованими процесами, забезпечуючи ефективну роботу виробництва. Вона - " "Кассандра вправно керує автомобілем, виявляючи високий рівень водійської майстерності. Вона - " "Мій чоловік втілює свої ідеї у власних творах, пишучи захоплюючі оповідання та книги. Він - " "Мій колишній однокласник ефективно підбадьорює та мобілізує громаду, виступаючи за ідеї. Він - " "Марія вивчає та вдосконалює методи сільськогосподарського виробництва, бережливо ставлячись до рослин. Вона - " "Мій брат захищає права та інтереси клієнтів у суді, професійно вирішуючи юридичні питання. Він - " "Кассандра відстоює ідеї анархії, агітуючи за вільні та незалежні структури суспільства. Вона - " "Мій чоловік є представником англійської національності, пишаючись своєю культурною спадщиною. Він - " "Марія Петрівна вивчає різноманітні аспекти людської культури та поведінки, розкриваючи таємниці соціального взаємодії. Вона - " "Мій колишній однокласник відомого вченого активно допомагає у проведенні досліджень та лабораторних робіт. Він - " "Мій брат вивчає космос та вчиться до майбутнього польоту у космос як астронавт. Він - " "Марія забезпечує нашу місцеву аптеку ліками та медичними засобами, дбаючи про здоров'я громади. Вона - "I get:
"Моя колишня однокласниця готується до вступу у виш, здібна та наполеглива. Вона - абітурієнтка." "Мій брат творить вражаючі образи, захоплюючи глядачів абстракцією. Він - художник." "Марія Петрівна керує автоматизованими процесами, забезпечуючи ефективну роботу виробництва. Вона - менеджер виробництва." "Кассандра вправно керує автомобілем, виявляючи високий рівень водійської майстерності. Вона - водій." "Мій чоловік втілює свої ідеї у власних творах, пишучи захоплюючі оповідання та книги. Він - письменник." "Мій колишній однокласник ефективно підбадьорює та мобілізує громаду, виступаючи за ідеї. Він - активіст." "Марія вивчає та вдосконалює методи сільськогосподарського виробництва, бережливо ставлячись до рослин. Вона - агроном." "Мій брат захищає права та інтереси клієнтів у суді, професійно вирішуючи юридичні питання. Він - адвокат." "Кассандра відстоює ідеї анархії, агітуючи за вільні та незалежні структури суспільства. Вона - анархістка." "Мій чоловік є представником англійської національності, пишаючись своєю культурною спадщиною. Він - англієць." "Марія Петрівна вивчає різноманітні аспекти людської культури та поведінки, розкриваючи таємниці соціального взаємодії. Вона - антропологиня." "Мій колишній однокласник відомого вченого активно допомагає у проведенні досліджень та лабораторних робіт. Він - асистент вченого." "Мій брат вивчає космос та вчиться до майбутнього польоту у космос як астронавт. Він - астронавт." "Марія забезпечує нашу місцеву аптеку ліками та медичними засобами, дбаючи про здоров'я громади. Вона - фармацевтка."Fazit
- Get a list of names, entities, roles etc., to make it interesting.
- Get it to make multiple descriptions of each profession
- Save as a data structure like:
profession: str descriptions_male: list[str] descriptions_female: list[str] valid_feminitives: list[str] valid_masc.: list[str] - Generate the individual training instances.
Playing with python
LLM library
LLM: A CLI utility and Python library for interacting with Large Language Models (for later: install the GPT4All plugin for local uncensored models)
Templates: Prompt templates - LLM
llm templates edit m-femprompt: > Наведи будь-ласка однозначні короткі дефініції цим професіям чи словам, так, щоб по ним було однозначно очевидно про яку саме професію йде мова. Зміни дефініції так, щоб вони стали фразами, де мова однозначно йде про жінку. Придумай різні варіанти жінок, про яких йде мова, умовно: моя сестра, моя жінка, колишня однокласниця, імена (Марія, Марія Петрівна, Кассандра). Але придумай і свої різноманітніші приклади. Уникай використання самого слова чи поняття у визначеннях. Уникай слів 'фахівецька' чи 'спеціалістка'. Наприклад: Актор: "Моя жінка виконує ролі на сцені чи екрані" Акушерка: "Марія Петрівна допомагає при пологах" Автор: "Я знаю дівчину, яка пише твори та книжки". Будь творчим. Але професія, про яку іде мова, має все рівно бути однозначно зрозумілою. Зроби це для всіх цих слів: $input> echo "водій, програміст, вчений" | llm -t m-fem Водій: "Моя сестра допомагає перевозити пасажирів та вантажі на автомобілі" Програміст: "Моя однокласниця створює програми та розробляє веб-сайти" Вчений: "Кассандра проводить дослідження і займається науковими відкриттями"Damn!
# Why three? > echo "водій, програміст, вчений" | llm -t m-fem -m 3.5-16k Водій: "Моя колишня однокласниця розвозить пасажирів на таксі" Водій: "Марія перевозить вантажі на великогабаритних автомобілях" Водій: "Кассандра працює на міському автобусному маршруті" Програміст: "Моя сестра створює програми та веб-сайти для клієнтів" Програміст: "Марія Петрівна програмує роботів та штучний інтелект" Програміст: "Кассандра розробляє мобільні додатки для смартфонів" Вчений: "Моя жінка проводить експерименти та дослідження в лабораторії" Вчений: "Марія займається науковим дослідженням у галузі генетики" Вчений: "Кассандра вивчає космос та проводить астрономічні спостереження" > echo "водій, програміст, вчений" | llm -t m-fem -m 4 Водій: "Моя сестра керує автобусом, вантажівкою або автомобілем." Або: "Моя дружина впевнено сідає за кермо і відповідально перевозить пасажирів." Або: "Ката наполягає, що її життєвою страстю є перевезення товарів на великі відстані." Програміст: "Моя колишня однокласниця говорить мовою Python, Java або C++, розробляючи програмне забезпечення." Або: "Моєму серцю неодмінно стане на шляху любов до жінок, що пишуть код і створюють додатки." ^CI got it, larger models interpret “definitions” etc. as an instruction to create multiple options for each word! Wow!
LangChain
COMPLETE_PROMPT: str = """ Наведи будь-ласка однозначні короткі дефініції цим професіям чи словам, так, щоб по ним було однозначно очевидно про яку саме професію йде мова. Зміни дефініції так, щоб вони стали фразами, де мова однозначно йде про жінку. Придумай різні варіанти жінок, про яких йде мова, умовно: моя сестра, моя жінка, колишня однокласниця, імена (Марія, Марія Петрівна, Кассандра). Але придумай і свої різноманітніші приклади. Уникай використання самого слова чи поняття у визначеннях. Уникай слів 'фахівецька' чи 'спеціалістка'. Наприклад: Актор: "Моя жінка виконує ролі на сцені чи екрані" Акушерка: "Марія Петрівна допомагає при пологах" Автор: "Я знаю дівчину, яка пише твори та книжки". Будь творчим. Але професія, про яку іде мова, має все рівно бути однозначно зрозумілою. """ FORMAT_INSTRUCTIONS = """ Формат виводу - JSON, по обʼєкту на кожну дефініцію. Обʼєкт виглядати таким чином: { "profession": "", "description": "" } Виводь тільки код JSON, без ніяких додаткових даних до чи після. """ prompt = PromptTemplate( template="{complete_prompt}\n{format_instructions}\n Професія, яку потрібно описати: {query}\n", input_variables=["query"], partial_variables={ "format_instructions": FORMAT_INSTRUCTIONS, "complete_prompt": COMPLETE_PROMPT, }, ) json_parser = SimpleJsonOutputParser() prompt_and_model = prompt | model | json_parser output = prompt_and_model.invoke({"query": "архітектор,програміст"})[{'description': ['Моя сестра працює в школі і навчає дітей', 'Дочка маминої подруги викладає у початковій ' 'школі'], 'profession': 'Вчителька'}, {'description': ['Моя сестра створює картини, які відображають ' 'абстрактні ідеї та почуття', 'Дівчина, яку я знаю, малює абстракціоністські ' 'полотна'], 'profession': 'абстракціоністка'}, {'description': ['Моя сестра вміє водити автомобіль', 'Дівчина знає всі тонкощі водіння автомобіля'], 'profession': 'автомобілістка'}, {'description': ['Моя сестра пише книги та статті', 'Дівчина, яку я знаю, створює літературні твори', 'Марія Петрівна є відомою письменницею'], 'profession': 'авторка'}, {'description': ['Моя сестра вивчає рослинництво та допомагає ' 'фермерам у вирощуванні культур', 'Дочка маминої подруги консультує селян щодо ' 'вибору добрив та захисту рослин'], 'profession': 'агрономка'}, {'description': ['Моя сестра захищає клієнтів у суді', 'Дочка маминої подруги працює в юридичній фірмі'], 'profession': 'адвокатка'}, {'description': ['Моя сестра бореться за відсутність влади та ' 'держави', 'Дівчина, яку я знаю, вірить у самоорганізацію ' 'суспільства без уряду'], 'profession': 'анархіст'}, {'description': ['Моя колишня однокласниця живе в Англії', 'Моя сестра вивчає англійську мову'], 'profession': 'англієць'}, {'description': ['Моя сестра вивчає культури та традиції різних ' 'народів', 'Дочка маминої подруги досліджує етнічні групи ' 'та їхні звичаї'], 'profession': 'антрополог'}, {'description': ['Моя сестра допомагає виконувати різні завдання ' 'на роботі', 'Дочка маминої подруги організовує робочий ' 'графік та зустрічі'], 'profession': 'асистентка'}, {'description': ['Моя сестра досліджує космос як астронавт', 'Дочка маминої подруги летить у космос як ' 'астронавт'], 'profession': 'астронавт'}, {'description': ['Моя сестра працює в аптеці та консультує ' 'пацієнтів з ліками', 'Дочка маминої подруги видає ліки в аптеці'], 'profession': 'аптекар'}, {'description': ['Моя сестра працює в школі та навчає дітей', 'Дочка маминої подруги викладає у початковій ' 'школі'], 'profession': 'Вчителька'}]These generate worse prompts:
COMPLETE_PROMPT: str = """Наведи будь-ласка {N_PROFS} однозначні короткі дефініції цій професії або слову, так, щоб по ним було однозначно очевидно про яку саме професію йде мова. Зроби два варіанта дефініцій: 1) Зміни дефініції так, щоб вони стали фразами, де мова однозначно йде про жінку. Придумай різні варіанти жінок, про яких йде мова, умовно: {WOMEN_VARIANTS}. Але придумай і свої різноманітніші приклади. 2) Те саме, але про чоловіків. Опис професії де мова йде про чоловіка. Уникай використання самого слова чи поняття у визначеннях. Уникай слів 'фахівецька' чи 'спеціалістка'. Наприклад: Актор: "Моя жінка виконує ролі на сцені чи екрані", "Мій чоловік виконує ролі на сцені чи екрані" Акушерка: "Марія Петрівна допомагає при пологах", "Валентин Петрович допомагає при пологах" Автор: "Я знаю дівчину, яка пише твори та книжки", "Я знаю хлопця, який пише твори та книжки" Будь творчим. Але професія, про яку іде мова, має все рівно бути однозначно зрозумілою. """ FORMAT_INSTRUCTIONS = """ Формат виводу - JSON. Обʼєкт виглядати таким чином: { "profession": "", "description": [ [description_female, description_male], [description_female, description_male], ] } В полі description список всіх згенерованих дефініцій, для кожної з якої надається пара жіночого опису і чоловічого. Виводь тільки код JSON, без ніяких додаткових даних до чи після. """Problems I found
- LangChain with its English-language description of the JSON schema didn’t work well for Ukrainian-language instructions, and I had to write them manually skipping the whole Schema part
Notes from 231010-1003 Masterarbeit Tagebuch
231010-1003 Masterarbeit Tagebuch#Feminitives task 231204-1642 Masterarbeit evaluation task new UA grammar and feminitives
- Adding male examples would be a really cool baseline
- slot filling is the keyword for this kind of task and it’s a solved problem
Existing stuff
-
Day 1798 (03 Dec 2023)
Masterarbeit eval task LMentry-static-UA
Context: 220120-1959 taskwarrior renaming work tasks from previous work
First notes
Just tested this: DAMN!
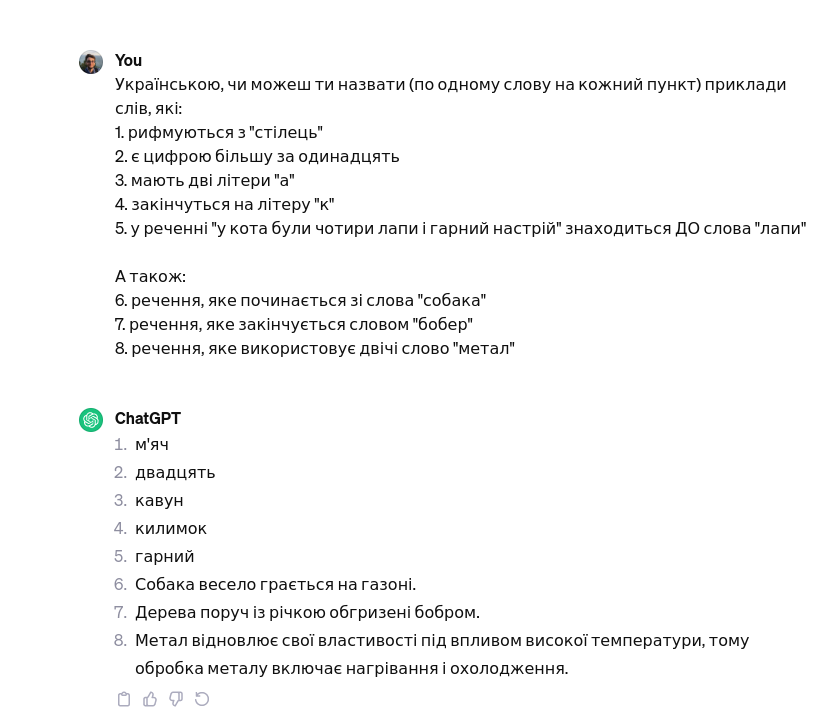
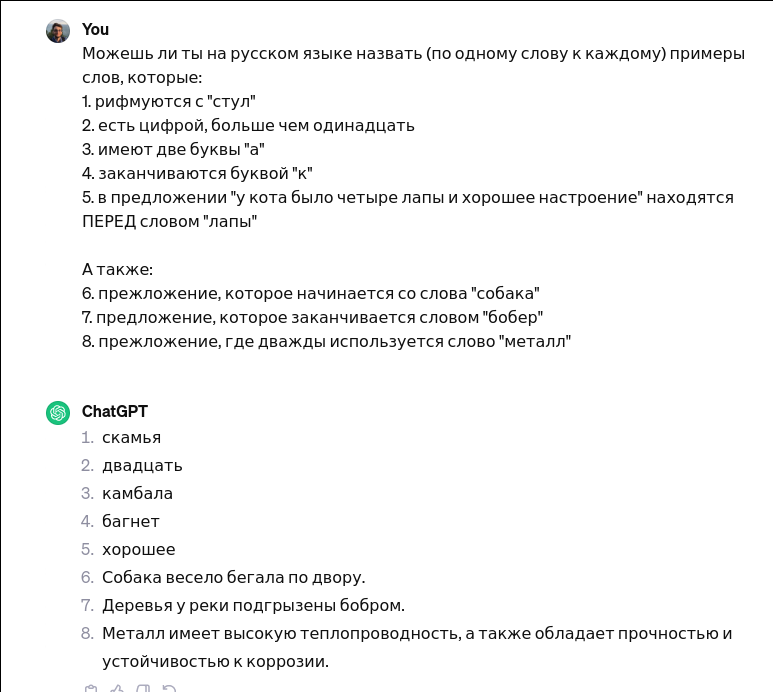
User Can you, in English, name one word for each of these tasks: 1. Rhymes with "chair" 2. Is a number larger than eleven 3. Has two letters "a" 4. Ends with the letter "k" 5. In the sentence "the cat had four paws and a good mood" is BEFORE the word "paws" Also: 6. A sentence that starts with the word "dogs" 7. A sentence that ends with the word "beaver" 8. A sentence that uses the word "metal" twicehttps://chat.openai.com/share/3fdfaf05-5c13-44eb-b73f-d66f33b73c59
lmentry/data/all_words_from_category.json at main · aviaefrat/lmentry
Not all of it needs code and regexes! lmentry/data/bigger_number.json at main · aviaefrat/lmentry
I can really do a small lite-lite subset containing only tasks that are evaluatable as a dataset.
LMentry-micro-UA
// minimal, micro, pico
Plan:
- go methodically through all of those task, divide them into regex and not regex, clone the code translate the prompts generate the dataset
Decision on 231010-1003 Masterarbeit Tagebuch#LMentry-micro-UA: doing a smaller version works!
LMentry-static-UA
Basics
Will contain only a subset of tasks, the ones not needing regex. They are surprisingly many.
The code will generate a json dataset for all tasks.
Implementation
Original task/code/paper analysis
- lmentry/resources at main · aviaefrat/lmentry has the JSONs with static words etc. used to generate the tasks
- lmentry/lmentry/predict.py at main · aviaefrat/lmentry contains the predicting code used to evaluate it using different kinds of models - I’ll need this later.
- tasks enumeration:
- lmentry/lmentry/constants.py at main · aviaefrat/lmentry list of all tested models
- static tasks (
won’timplement, completed):Sentence containing wordSentence not containing wordWord containing letterWord not containing letter- Most associated word
- Least associated word
- Any words from category
- All words from category
- First alphabetically
- More letters
- Less letters
- Bigger number
- Smaller number
- Rhyming word
HomophonesWon’t do because eight/ate won’t work in ukr- Word after in sentence
- Word before in sentence
Sentence starting with wordSentence ending with wordWord starting with letterWord ending with letter- First word of the sentence
- Last word of the sentence
- First letter of the word
- Last letter of the word
Distractors
- order of words
- template content
- adj tasks VS arg. content
My changes
- I’d like to have words separated by:
- frequency
- length,
- … and maybe do cool analyses based on that
- (DONE) in addition to first/last letter/word in word/sentence, add arbitrary “what’s the fourth letter in the word ‘word’?”
- Longer/shorter words: add same length as option
My code bits
- I have to write it in a way that I can analyze it for stability wrt morphology etc. later
Ukrainian numerals creation
Problem: ‘1’ -> один/перший/(на) першому (місці)/першою
Existing solutions:
- pymorphy can inflect existing words but needs
- savoirfairelinux/num2words: Modules to convert numbers to words. 42 –> forty-two can’t do ordinals+case
Created my own! TODO document
TODO https://webpen.com.ua/pages/Morphology_and_spelling/numerals_declination.html
More tagsets fun
Parse(word='перша', tag=OpencorporaTag('ADJF,compb femn,nomn'), normal_form='перший', score=1.0, methods_stack=((DictionaryAnalyzer(), 'перша', 76, 9),))compbNothing in docu, found it only in the Ukr dict converter tagsets mapping: LT2OpenCorpora/lt2opencorpora/mapping.csv at master · dchaplinsky/LT2OpenCorporaI assume it should get converted tocompbut doesn’t - yet another future bug report to pymorphy4EDIT 2025-06-03: Casey Jones (caseyjones.us) emailed me with a much better explanation! Quoting (bold text mine):
The link you provide there to LT2OpenCorpora/lt2opencorpora/mapping.csv was helpful for me.
It lists:
- compb: базова форма
- compr: порівняльна форма
- super: найвища форма
You wrote:
I assume it should get converted to comp but doesn’t - yet another future bug report to pymorphy4
But I don’t think so. My interpretation of the above is that “compb” stands for “comparable BASE form”, which makes sense once you figure it out, but that should definitely be better documented. And it’s weird then that
comprandsuperaren’t consistently namedcompcandcompsrespectively, but whatever, I guess, if only people can figure out what they mean.Even more tagsets fun
pymorphy2 doesn’t add the
singtag for Ukrainian singular words. Then any inflection that deals with number fails.Same issue I had in 231024-1704 Master thesis task CBT
Found a way around it:
@staticmethod def _add_sing_to_parse(parse: Parse) -> Parse: """ pymorphy sometimes doesn't add singular for ukrainian (and fails when needs to inflect it to plural etc.) this creates a new Parse with that added. """ if parse.tag.number is not None: return parse new_tag_str = str(parse.tag) new_tag_str+=",sing" new_tag = parse._morph.TagClass(tag=new_tag_str) new_best_parse = Parse(word=parse.word, tag=new_tag, normal_form=parse.normal_form, score=parse.score, methods_stack=parse.methods_stack) new_best_parse._morph=parse._morph return new_best_parse # Not needed for LMentry, but I'll need it for CBT anyway... @staticmethod def _make_agree_with_number(parse: Parse, n: int)->Parse: grams = parse.tag.numeral_agreement_grammemes(n) new_parse = Numbers._inflect(parse=parse, new_grammemes=grams) return new_parseparse._morphis the Morph.. instance, without one added inflections of that Parse fail.TagClassfollows the recommendations of the docu2 that say better it than a newOpencorporaTag, even though both return the same class.
Notes by task
Taxonomy
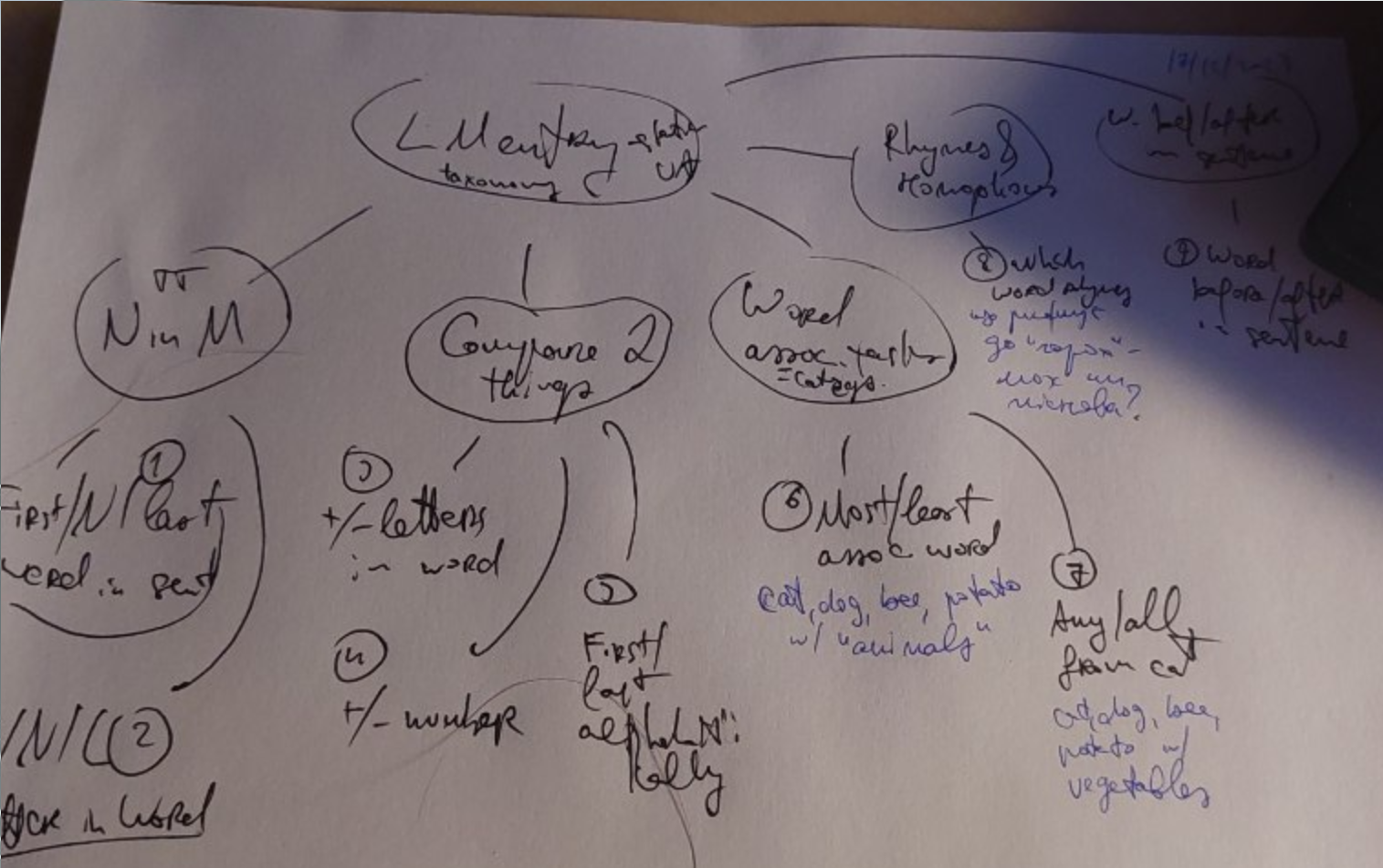 +
+Comparing two things
Words of different lengths, alphabet order of words, etc.
Main relationship is
kind=less|more, wherelessmeans “word closer to beginning of the alphabet”, “smaller number”, “word with fewer letters” etc.,moreis the opposite.Alphabet order of words
- (DONE) which word is closer to beginning of alphabet
- Are these words in alphabet order?
Which word is longer
TODO Which number is bigger
- use the one-million bits and add to the text that this is why I needed to care about agreemnet
- do comparisons of entities! one box has a million pencils, the other has five hundred thousand. Which has more pencils?
GPT4 agreement issues
https://chat.openai.com/share/b52baed7-5d56-4823-af3e-75a4ea8d5b8c: 1.5 errors, but I’m not sure myself about the fourth one.
LIST = [ "Яке слово стоїть ближче до початку алфавіту: '{t1}' чи '{t2}'?", "Що є далі в алфавіті: '{t1}' чи '{t2}'?", "Між '{t1}' та '{t2}', яке слово розташоване ближче до кінця алфавіту?", # TODO - в алфавіті? "У порівнянні '{t1}' і '{t2}', яке слово знаходиться ближче до A в алфавіті?", # ChatGPT used wrong відмінок внизу: # "Визначте, яке з цих слів '{t1}' або '{t2}' знаходиться далі по алфавіті?", ]HF Dataset
I want a ds with multiple configs.
Base patterns
- HA! Lmentry explicitly lists base patterns: lmentry/lmentry/scorers/first_letter_scorer.py at main · aviaefrat/lmentry
starts = "(starts|begins)" base_patterns = [ rf"The first letter is {answer}", rf"The first letter {of} {word} is {answer}", rf"{answer} is the first letter {of} {word}", rf"{word} {starts} with {answer}", rf"The letter that {word} {starts} with is {answer}", rf"{answer} is the starting letter {of} {word}", rf"{word}: {answer}", rf"First letter: {answer}", ]For more: lmentry/lmentry/scorers/more_letters_scorer.py at main · aviaefrat/lmentry
Looking for example sentences
- spacy example sentences
- political ones from UP!
- implemented
- for words, I really should use some normal dictionary.
Assoc. words and resources
Another dictionary I found: slavkaa/ukraine_dictionary: Словник слів українською (слова, словоформи, синтаксичні данні, літературні джерела)
- Excel_word_v10.xslx
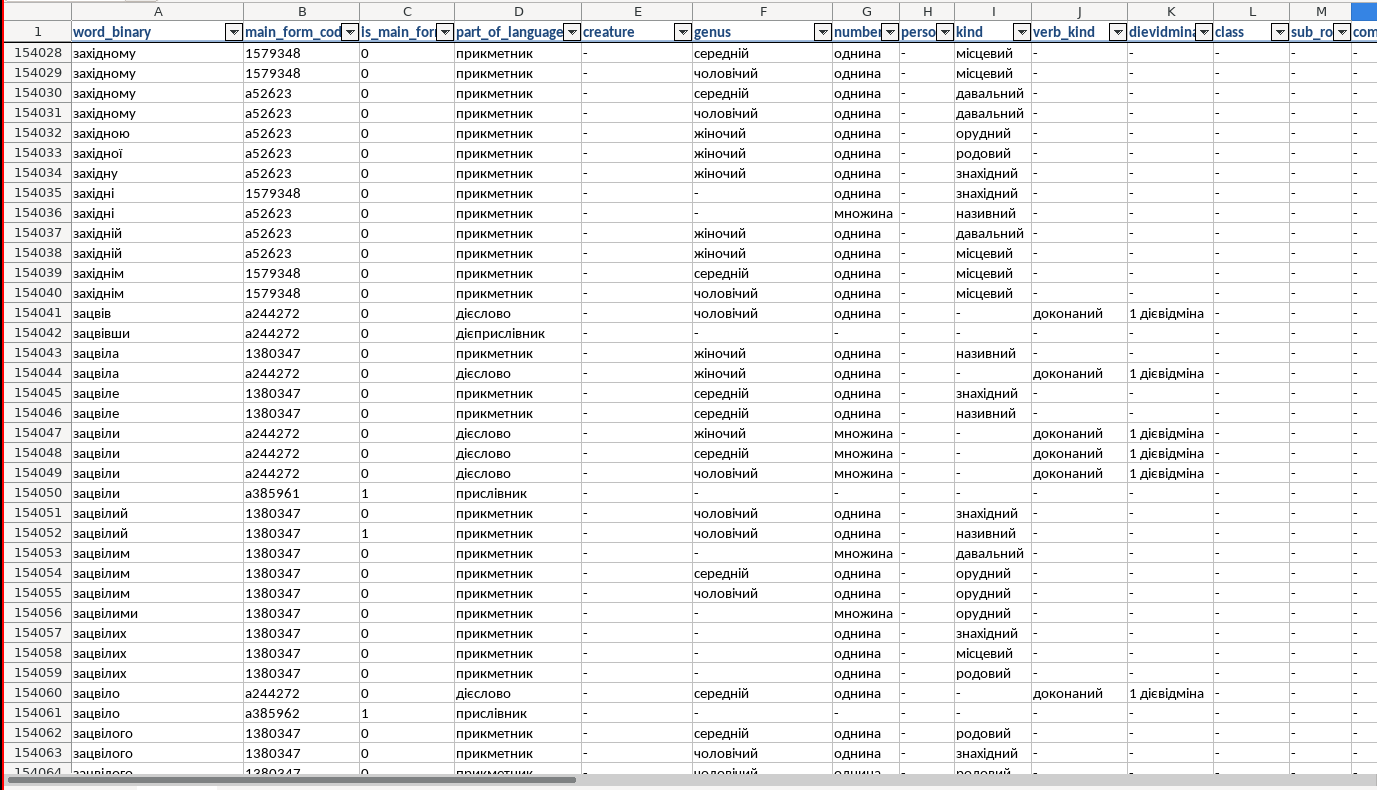
- sql as well
- a lot of columns
Next tasks
- List
- Most associated word
- Least associated word
- Any words from category
- All words from category
All basically need words and their categories. E.g. Animals: dog/cat/racoon
I wonder how many different categories I’d need
Ah, the O.G. benchmark has 5 categories: lmentry/resources/nouns-by-category.json at main · aviaefrat/lmentry
Anyway - I can find no easy dictionary about this.
options:
Wordnet
- Other Wordnet: olgakanishcheva/WordNet-Affect-UKR: WordNet-Affect (http://wndomains.fbk.eu/wnaffect.html) is an extension of WordNet Domains, including a subset of synsets suitable to represent affective concepts correlated with affective words.
- multi-lang incl. Ukr
- emotions only
- txts are tab-separated, colums are lists of words in column-language
- WordNet — someone started it for UKR!
- wordnet/resources/wn-ua-2015 at main · lang-uk/wordnet
- wordnet/assumptions.md at main · lang-uk/wordnet this is so cool!
- wordnet/notebooks/wn-translation-analysis-khrystyna.md at main · lang-uk/wordnet 10/10 would love to work on/at/with that/them
- 2015
- all-dict-entries: definitions
- all-in-one-file: relationships between these definitions
- wordnet/resources/wn-ua-2015 at main · lang-uk/wordnet
- WordNet - Wikipedia:
- hypernym: Y is a hypernym of X if every X is a (kind of) Y (canine is a hypernym of dog)
- hyponym: Y is a hyponym of X if every Y is a (kind of) X (dog is a hyponym of canine)
- holonym: Y is a holonym of X if X is a part of Y (building is a holonym of window)
- meronym: Y is a meronym of X if Y is a part of X (window is a meronym of building)
for all-in-one:
> grep -o "_\(.*\)(" all-in-one-file.txt | sort | uniq -c 49 _action( 8 _action-and-condition( 58 _holonym( 177 _hyponym( 43 _meronym( 12 _related( 51 _sister( 102 _synonym(looking through it it’s sadly prolly too small
2009’s hyponym.txt is nice and much more easy to parse.
GPT
Ideas: WordNet Search - 3.1 Ask it to give me a list of:
- emotions
- professions
- sciences
- body parts
- animals
- times (dow, months, evening, etc.)
- sports It suggests also
- musical instruments
- dishes
- clothing
-
<_(@bm_lmentry) “LMentry: A language model benchmark of elementary language tasks” (2022) / Avia Efrat, Or Honovich, Omer Levy: z / / 10.48550/ARXIV.2211.02069 _> ↩︎
-
API Reference (auto-generated) — Морфологический анализатор pymorphy2 ↩︎
-
Day 1796 (01 Dec 2023)
Pip can easily install packages from github
Created pchr8/pymorphy-spacy-disambiguation: A package that picks the correct pymorphy2 morphology analysis based on morphology data from spacy to easily include it in my current master thesis code.
Later on releases pypi etc., but for now I just wanted to install it from github, and wanted to know what’s the minimum I can do to make it installable from github through pip.
To my surprise,
pip install git+https://github.com/pchr8/pymorphy-spacy-disambiguationworked as-is! Apparently pip is smart enough to parse the poetry project and run the correct commands.poetry add git+https://github.com/pchr8/pymorphy-spacy-disambiguationworks just as well.
Otherwise, locally:
poetry buildcreates a
./distdirectory with the package as installable/shareable files.Also, TIL:
poetry show poetry show --tree --why coloramashow a neat colorful tree of package dependencies in the project.
How to read and write a paper according to hackernews
- How to Read a Paper [pdf] | Hacker News (pdf)
-
I give all my doctoral students a copy of the following great paper (and I’ve used a variant of the check list at the end for years - avoids errors when working on multiple papers with multiple international teams in parallel) http://www-mech.eng.cam.ac.uk/mmd/ashby-paper-V6.pdf
-
I’ll write here the main points from each of the linked PDF, copyright belongs to the original authors ofc.
How to Write a Paper
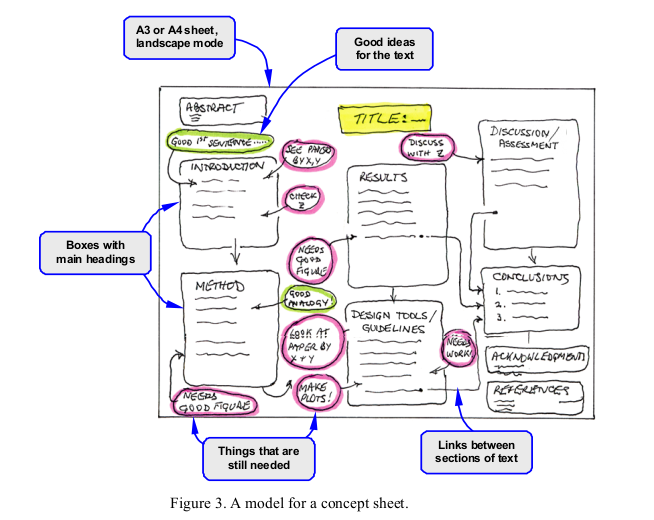
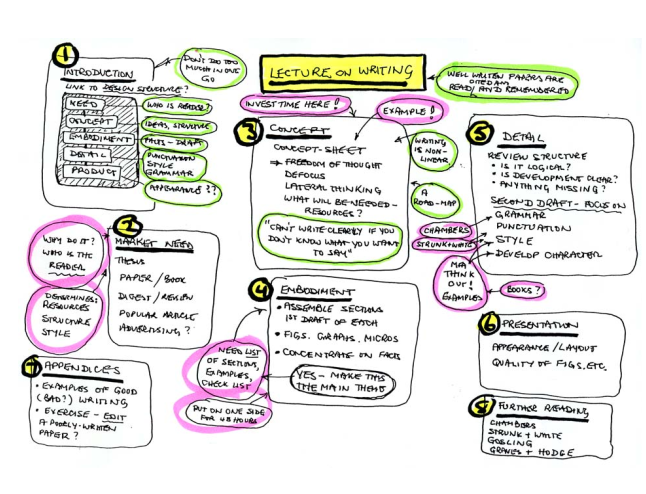
How to Write a Paper Mike Ashby Engineering Department, University of Cambridge, Cambridge 6 rd Edition, April 2005This brief manual gives guidance in writing a paper about your research. Most of the advice applies equally to your thesis or to writing a research proposal.
This is based on 2016 version of the paper, more are here: https://news.ycombinator.com/item?id=38446418#38449638 with the link to the 2016 version being https://web.archive.org/web/20220615001635/http://blizzard.cs.uwaterloo.ca/keshav/home/Papers/data/07/paper-reading.pdf
- The design
- The market need - what is the purpose? Who will read it? How will it be used?
- Thesis / paper / research-proposal:
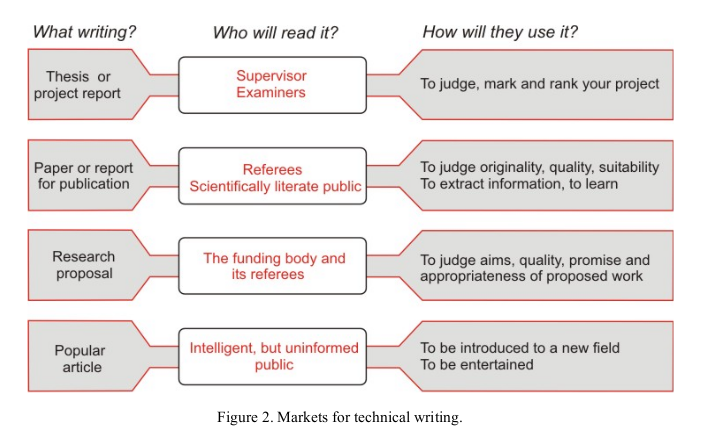
- Thesis / paper / research-proposal:
- The market need - what is the purpose? Who will read it? How will it be used?
- Concept
-
When you can’t write, it is because you don’t know what you want to say. The first job is to structure your thinking.
- A3 paper where you draw things:
-
-
Don’t yet think of style, neatness or anything else. Just add, at the appropriate place on the sheet, your thoughts.
-
- Embodiement
- the first draft
- the PDF lists random bits about each sections, like abstract / introduction / …
- Introduction:
- What is the problem and why is it interesting?
- Who are the main contributors?
- What did they do?
- What novel thing will you reveal?
- Method
- ‘just say what you did, succinctly’
- Results
- Same; also succinctly, without interpretation etc.
- …
- Appendices:
- essential material that would interrupt the flow of the main text
- Grammar!
-

- That VS which!

-
- Punctuation:
- really interested and itemized
- Dashes: “The dash sets off parenthetic material that results in a break in continuity in a sentence. [..] A dash can lead to an upshot, a final summary word or statement, and give emphasis:”
- Parentheses—literally: putting-asides—embrace material of all sorts, and help structure scientific writing. But do not let them take over, clouding the meaning of the sentence with too many asides.
- Italics: the best of the three ways (with bold and underline) to emphasize stuff in scientific writing.
- Brackets are used to indicate editorial comments or words inserted as explanation:
[continued on p. 62],[see footnote].
- Style
- Be clear. Use simple language,familiar words, etc.
- Design: Remember who you are writing for. Tell them what they want to know, not what they know already or do not want to know.
- Define everything
- Avoid cliches; avoid empty words
-
Avoid clichés (standard formalised phrases): they are corpses devoid of the vitality which makes meaning spring from the page
-

-
- Do not overstate, over emphasise or apologise:
не верь, не бойся не проси
- Avoid being patronising, condescending or eccentric
- Good first sentence:
- Openings such as: It is widely accepted that X (your topic) is important … has the reader yawning before you’ve started.
- At the end it has examles of effective and ineffective writing
- At the very end it has this:
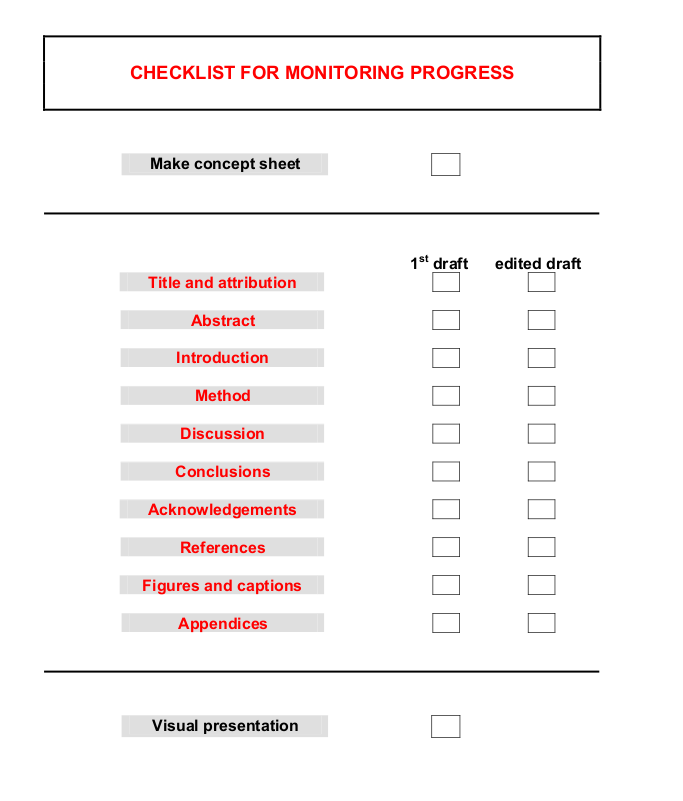
How to read a paper
How to Read a Paper S. Keshav David R. Cheriton School of Computer Science, University of Waterloo Waterloo, ON, Canada keshav@uwaterloo.cahttp://ccr.sigcomm.org/online/files/p83-keshavA.pdf
- Three passes of varying levels of thoroughness
- Literature survery:
- also three steps:
- find recent papers in the area through google scholar etc.
- find top conferences
- look through their recent conference proceedings
- also three steps:
- How to Read a Paper [pdf] | Hacker News (pdf)
-
Day 1792 (27 Nov 2023)
pytest and lru_cache
I have a pytest of a function that uses python
@lru_cache:cacheinfo = gbif_get_taxonomy_id.cache_info() assert cacheinfo.hits == 1 assert cacheinfo.misses == 2LRU cache gets preserved among test runs, breaking independence and making such bits fail.
Enter pytest-antilru · PyPI which resets the LRU cache between test runs. Installing it as a python package is all there’s to ite.
Passing booleans to python argparse as str
Needed argparse to accept yes/no decisions, should have been used inside a dockerfile that doesn’t have if/else logic, and all solutions except getting a parameter that accepts string like true/false seemed ugly.
The standard linux
--do-thingand--no-do-thingwere also impossible to do within Docker, if I want to use an env. variable etc., unless I literally set them to--do-thingwhich is a mess for many reasons.I had 40 tabs open because apparently this is not a solved problem, and all ideas I had felt ugly.
How do I convert strings to bools in a good way? (
boolalone is not an option becausebool('False')etc.)Basic
if value=="true"would work, but maybe let’s support other things as a bonus because why not.My first thought was to see what YAML does, but then I found the deprecated in 3.12
distutils.util.strtobool: 9. API Reference — Python 3.9.17 documentationIt converts y,yes,t,true,on,1 / n,no,f,false,off,0 into boolean
True/False.The code, the only reason it’s a separate function (and not a lambda inside the
type=parameter) was because I wanted a custom ValueError and to add the warning for deprecation, as if Python would let me forget. An one-liner was absolutely possible here as well.def _str_to_bool(x: str): """Converts value to a boolean. Currently uses (the rules from) distutils.util.strtobool: (https://docs.python.org/3.9/distutils/apiref.html#distutils.util.strtobool) True values are y, yes, t, true, on and 1 False values are n, no, f, false, off and 0 ValueError otherwise. ! distutils.util.strtobool is deprecated in python 3.12 TODO solve it differently by then Args: value (str): value """ try: res = bool(strtobool(str(x).strip())) except ValueError as e: logger.error( f"Invalid str-to-bool value '{x}'. Valid values are: y,yes,t,true,on,1 / n,no,f,false,off,0." ) raise e return res # inside argparse parser.add_argument( "--skip-cert-check", help="Whether to skip a cert check (%(default)s)", type=_str_to_bool, default=SKIP_CERT_CHECK, )This allows:
- sane human-readable default values specified elsewhere
- use inside Dockerfiles and Rancher configmaps etc. where you just set it to a plaintext value
- no if/else bits for
--no-do-thingflags
distutilsis deprecated in 3.12 though :(
YAML is known for it’s bool handling: Boolean Language-Independent Type for YAML™ Version 1.1.
Regexp:
y|Y|yes|Yes|YES|n|N|no|No|NO |true|True|TRUE|false|False|FALSE |on|On|ON|off|Off|OFF`I don’t like it and think it creates more issues than it solves, e.g. the “Norway problem” (211020-1304 YAML Norway issues), but for CLI I think that’s okay enough.
Rancher secrets and config maps
Using Kubernetes envFrom for environment variables describes how to get env variables from config map or secret, copying here:
##################### ### deployment.yml ##################### # Use envFrom to load Secrets and ConfigMaps into environment variables apiVersion: apps/v1beta2 kind: Deployment metadata: name: mans-not-hot labels: app: mans-not-hot spec: replicas: 1 selector: matchLabels: app: mans-not-hot template: metadata: labels: app: mans-not-hot spec: containers: - name: app image: gcr.io/mans-not-hot/app:bed1f9d4 imagePullPolicy: Always ports: - containerPort: 80 envFrom: - configMapRef: name: env-configmap - secretRef: name: env-secrets ##################### ### env-configmap.yml ##################### # Use config map for not-secret configuration data apiVersion: v1 kind: ConfigMap metadata: name: env-configmap data: APP_NAME: Mans Not Hot APP_ENV: production ##################### ### env-secrets.yml ##################### # Use secrets for things which are actually secret like API keys, credentials, etc # Base64 encode the values stored in a Kubernetes Secret: $ pbpaste | base64 | pbcopy # The --decode flag is convenient: $ pbpaste | base64 --decode apiVersion: v1 kind: Secret metadata: name: env-secrets type: Opaque data: DB_PASSWORD: cDZbUGVXeU5e0ZW REDIS_PASSWORD: AAZbUGVXeU5e0ZB @caiquecastroThis is neater than what I used before, listing literally all of them:
spec: containers: - name: name image: image env: - name: BUCKET_NAME valueFrom: configMapKeyRef: name: some-config key: BUCKET_NAME
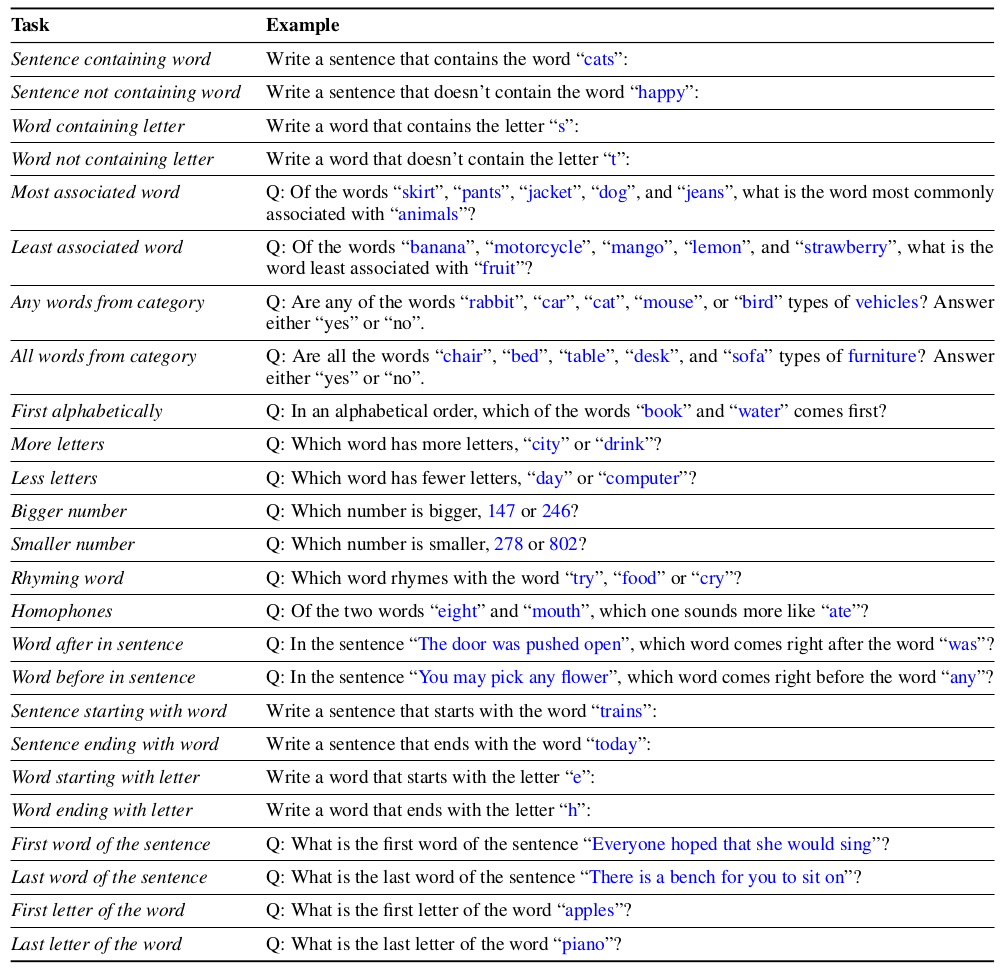 from the paper
from the paper